In 1994, a Swiss biologist named Pascal Gagneux began a Ph.D. program in zoology. His research plan was to stalk populations of wild chimpanzees in Cote d’Ivoire and Mali. Specifically, Gagneux was in search of chimp nests in forest trees. After he figured out where the chimps slept, he’d wait until they left during the day to search for food before climbing trees to collect their hair.
Gagneux’s aim was to document genetic diversity in our nearest, dearest, and critically endangered relatives. In order to see what made one chimp different from another, and one population of chimps different from the next, he needed to be able to take that hair he’d collected, extract DNA from its follicle, and quickly multiply the DNA. If Gagneux had embarked on his quest just a few years earlier, that crucial last step, the multiplying, wouldn’t have been possible. But thanks to a then-new scientific technique—and a fortuitous discovery in the hot springs of Yellowstone National Park two decades earlier—Gagneux was able to learn that chimpanzees have much more genetic diversity than humans. “There’s more diversity in one social group of chimpanzees than in all 7 billion humans on the planet,” he said.

Because the numbers of wild chimps are rapidly dwindling, that knowledge might have been completely lost were it not for the conveniently timed invention of the polymerase chain reaction, or PCR, a process that allows scientists to take a single scrap of DNA and churn out copy after copy with all the ease of stuffing paper into a Xerox. PCR, introduced in 1983, made Gagneux’s work possible.
In fact, PCR is one of the great technical achievements of our time. Many of the mapping techniques used in the Human Genome Project relied on PCR, and it’s helped do everything from identify a previously unknown bacteria that can cause a more-serious form of Lyme disease to aid doctors in screening patients for hereditary diseases like Tay-Sachs and cystic fibrosis. Its inventor, Kary Mullis, won the Nobel Prize for Chemistry in 1993.
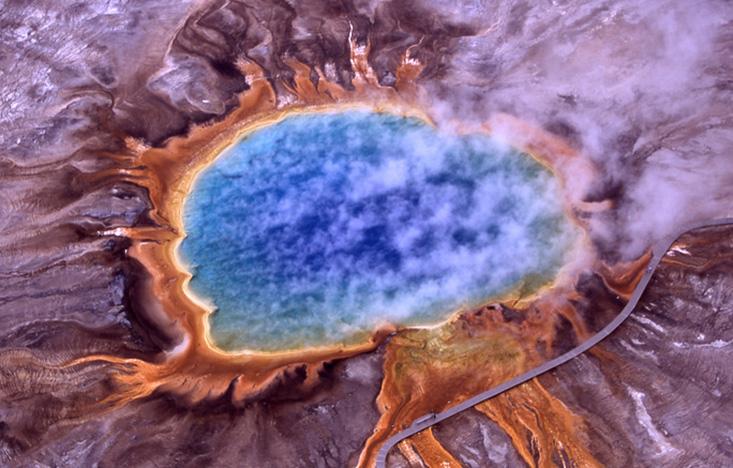
But the success of PCR and its standing as one of the most important scientific advances in the history of molecular biology depends upon the discovery, in the mid-1960s, of a pinkish-orange squiggle of a bacterium in a hot spring at Yellowstone National Park. The two men who discovered this bacterium didn’t go searching for something that could be used for DNA replication—or anything else, for that matter. They were just curious. And that curiosity just happened to change the world.
T. aquaticus would help spark a genetic revolution more than a decade later.
In the mid-1960s, when Hudson Freeze was an undergrad at Indiana University, he was, he said, likely the only guy in his dorm who didn’t smoke dope. “I was a classic nerd,” he said. (He even worked in one of the university’s labs while in high school.) Despite his interest in science, he describes himself as drifting through life. One afternoon, he saw a lecture by a professor named Thomas Brock that intrigued him.
Brock’s research centered on microbial ecology: He was interested in how photosynthetic microbes worked outside of a laboratory setting and how photosynthesis changed with temperature. Hot springs, with their variety of relatively stable temperature ranges, seemed like a great place to do that.
So in the summer of 1966, armed with a grant from the National Science Foundation, Brock and Freeze set out for Yellowstone to collect bacteria that thrives in hot springs. It was the first time Freeze had ever left Indiana. He spent the summer gathering, grinding, and extracting bacteria samples to ship back to Brock’s lab. That fall, Brock paid Freeze $2 an hour to put the samples from the hot springs in culture media, at different temperatures, to see what they would grow. One day, in a sample set to 176 degrees Fahrenheit (80 degrees Celsius)—a temperature at which Brock, Freeze, and the rest of the world assumed that nothing could survive—Freeze found the pink filaments of a previously unknown bacterium that Brock would later name Thermus aquaticus—literally, “hot water.” Brock would go on to find the bacterium in lots of different hot water environments—including the University of Indiana’s own plumbing system.
Freeze remembers feeling disappointed with the new creature’s name. “That’s lousy. Hot water? You can be more creative than that.” But he finished his work for Brock, graduated, and went on to another school to work on his Ph.D. Beyond the curiosity of its ability to survive at such high temperatures, he didn’t see much use for the bacterium he’d helped discover. “Some people from Cincinnati who worked for Proctor and Gamble came to see if there was any use for the organism. I think they wanted to put it in Tide,” he said. “But I didn’t think it would amount to anything. I just thought it was cute.”
After Freeze left Indiana, his initial sample of T. aquaticus, taken on Sept. 5, 1966 from Yellowstone’s Mushroom Spring and designated YT-1, went to the American Type Culture Collection in Washington, D.C. And that was where it would help spark a genetic revolution more than a decade later.
You can exponentially multiply the copies of your target DNA. In a matter of hours, you can create millions.
In the early 1980s, many researchers were looking for ways to make more copies of DNA faster. Say you have a drop of blood left at a crime scene and you want to see if the DNA matches that of a suspect. DNA is fragile stuff and that drop of blood probably won’t contain enough, on its own, to allow you to sequence it and make a match. But if you can take the DNA and make many, many copies of it, you’ll have enough to work with. Think about a small animal, like an ant. You might not notice just one ant—but if there were 1,000, they’d be a lot more visible.
The polymerase chain reaction makes copies of DNA and it doesn’t, strictly speaking, need T. acquaticus in order to work. Instead, what it needs is a polymerase, a type of enzyme involved in building and repairing DNA. Scientists take DNA from an organism that contains a segment of DNA they want to copy, and mix it with polymerase, a bunch of loose nucleotides to build DNA from, and short strands of nucleotides, called primers, that show the polymerase where to start building and where to stop. Then, it’s just a matter of heating and cooling. Heat the mixture up, and the DNA separates into two strands. Cool it down and the primers latch on. Heat it up again, and the polymerases go to work, turning loose nucleotides into DNA copies. If you run the cycles of heating and cooling over and over and over, you can exponentially multiply the copies of your target DNA. One becomes two. Two becomes four. Four becomes eight. In a matter of hours, you can create millions.
That was the basic concept that Mullis came up with, but getting it to work in a commercial way was a group effort. Norman Arnheim, a scientist who, together with Henry Erlich, also worked on the early development of PCR, says that the initial system was clunky and fussy. Heat deactivated the polymerase. So, in every cycle, scientists would have to open each test tube up and add more polymerase, at just the right temperature, so it could work but not die. “You’d have to do this forever. It was excruciatingly painful,” he says.
To get around the problem, Mullis’ team started looking for a polymerase that could survive under high heat. They found it in Freeze and Brock’s YT-1 sample. In 1989, Science magazine named the polymerase from T. aquaticus “Molecule of the Year.”
And that was when Freeze found out what had happened with his old discovery. The rights to the PCR technique sold to Hoffman-LaRoche Pharmaceuticals for some $300 million, but T. aquaticus itself was never patented. Most scientists who do basic research don’t get rich off it. Turning a profit was never their goal. “It doesn’t work that way in science,” Freeze said. Instead, they’re out to further human knowledge and make that knowledge available to others. Without them, the inventions that do make money could never happen.
Today, Freeze is the director of the human genetics program at the Sanford Burnham Prebys Medical Discovery Institute in La Jolla, California, making him one of the millions of scientists who uses PCR almost every day. “Here I am using the fruits of this discovery and I’m actually improving lives with it,” he said. “That makes the whole project, and by that, I mean my life in science, really meaningful.”
Gagneux, the chimp geneticist, is one of Freeze’s colleagues and considers the older man a giant in his field. Despite this, Gagneaux was surprised to learn, years after they met, about the contribution Freeze had made to science. That’s the other thing about basic science—not only does it not make scientists rich, it often doesn’t even make them famous. Basic science is necessary, but it’s also frequently overlooked.
In the summer of 2014, on a family vacation to Yellowstone, Gagneux went searching for the hot spring where T. aquaticus was discovered. The area was beautiful, he said, all these bubbling pools in a riot of colors, but also quiet and anonymous. There’s no sign alerting visitors about its history. “I was happy to think I saw that pool,” Gagneux said. “I sent Hud an email and told him, ‘I think I found it.’ ”
Maggie Koerth-Baker is the senior science reporter for FiveThirtyEight.com. Her work has appeared in The New York Times Magazine, The Atlantic, and other publications.
This story was commissioned by the Science Philanthropy Alliance as part of its Science to Society series, illustrating the importance of basic scientific research.