Is the natural world creative? Just take a look around it. Look at the brilliant plumage of tropical birds, the diverse pattern and shape of leaves, the cunning stratagems of microbes, the dazzling profusion of climbing, crawling, flying, swimming things. Look at the “grandeur” of life, the “endless forms most beautiful and most wonderful,” as Darwin put it. Isn’t that enough to persuade you?
Ah, but isn’t all this wonder simply the product of the blind fumbling of Darwinian evolution, that mindless machine which takes random variation and sieves it by natural selection? Well, not quite. You don’t have to be a benighted creationist, nor even a believer in divine providence, to argue that Darwin’s astonishing theory doesn’t fully explain why nature is so marvelously, endlessly inventive. “Darwin’s theory surely is the most important intellectual achievement of his time, perhaps of all time,” says evolutionary biologist Andreas Wagner of the University of Zurich. “But the biggest mystery about evolution eluded his theory. And he couldn’t even get close to solving it.”
What Wagner is talking about is how evolution innovates: as he puts it, “how the living world creates.” Natural selection supplies an incredibly powerful way of pruning variation into effective solutions to the challenges of the environment. But it can’t explain where all that variation came from. As the biologist Hugo de Vries wrote in 1905, “natural selection may explain the survival of the fittest, but it cannot explain the arrival of the fittest.” Over the past several years, Wagner and a handful of others have been starting to understand the origins of evolutionary innovation. Thanks to their findings so far, we can now see not only how Darwinian evolution works but why it works: what makes it possible.
A popular misconception is that all it takes for evolution to do something new is a random mutation of a gene—a mistake made as the gene is copied from one generation to the next, say. Most such mutations make things worse—the trait encoded by the gene is less effective for survival—and some are simply fatal. But once in a blue moon (the argument goes) a mutation will enhance the trait, and the greater survival prospects of the lucky recipient will spread that beneficial mutation through the population.
The trouble is that traits don’t in general map so neatly onto genes: They arise from interactions between many genes that regulate one another’s activity in complex networks, or “gene circuits.” No matter, you might think: Evolution has plenty of time, and it will find the “good” gene circuits eventually. But the math says otherwise.
Take, for example, the discovery within the field of evolutionary developmental biology that the different body plans of many complex organisms, including us, arise not from different genes but from different networks of gene interaction and expression in the same basic circuit, called the Hox gene circuit. To get from a snake to a human, you don’t need a bunch of completely different genes, but just a different pattern of wiring in essentially the same kind of Hox gene circuit. For these two vertebrates there are around 40 genes in the circuit. If you take account of the different ways that these genes might regulate one another (for example, by activation or suppression), you find that the number of possible circuits is more than 10700. That’s a lot, lot more than the number of fundamental particles in the observable universe. What, then, are the chances of evolution finding its way blindly to the viable “snake” or “human” traits (or phenotypes) for the Hox gene circuit? How on earth did evolution manage to rewire the Hox network of a Cambrian fish to create us?
Exactly which genes you have may not matter so much (within reason), because the job they do is more a property of the network in which they are embedded.
The galaxy of gene circuits isn’t the only mind-bogglingly immense terrain that evolution has to navigate in order to innovate. The same issues apply, for example, to metabolic networks. Organisms have had to find ways of getting their energy from whatever fuel happens to be on hand—typically the metabolisms of microorganisms run on compounds such as glucose, ethanol, or citrate. Ideally, their metabolic machinery of enzymes would run on more than just one of these, so that they’d have more options for survival. But how easy is it to adapt to other fuels? Even for a relatively small list of common metabolic fuels, the number of possible metabolisms of this sort is again astronomical.
The same explosion of combinatorial options happens for proteins, which are molecules made up of many tens to hundreds of amino acids bound together in chains and folded up into particular molecular shapes. There are 20 different amino acids found in natural proteins, and for proteins just 100 amino acids long (which are small ones) the number of permutations is 10130. Yet the 4 billion years of evolution so far have provided only enough time to create around 1050 different proteins. So how on earth has it managed to find ones that work?
Finally there’s RNA. This molecule, once considered the mere go-between in converting a sequence of chemical building blocks in a gene (called base pairs) into a sequence of amino acids in a protein that catalyses a biochemical reaction, is now afforded much more status. For one thing, because RNA molecules can imitate both genes, by encoding information in a sequence, and proteins, by folding up into shapes that act as chemical catalysts, RNA is the best candidate for the multi-tasking molecules that orchestrated the earliest life. What’s more, we now know that RNA molecules are important regulators of gene activity: Some genes interact with others via the RNA molecules they encode, which can act as switches to turn the expression of other genes up or down. Yet evolutionary innovation with RNA also faces an absurd profusion of combinatorial choices.
In all these cases the questions are the same, and point to, as Wagner puts it, a “dirty secret” behind the success of the so-called modern synthesis of Darwinian evolutionary theory and genetics. How does evolution find workable solutions when it lacks the means to explore even a small fraction of the options? And how does evolution find its way from an existing solution to a viable new one—how does it create? The answer is, at least in part, a simple one: It’s easier than it looks. But only because the landscape that the evolutionary process explores has a remarkable structure, and one that neither Darwin nor his successors who merged Darwinism with genetics had anticipated.
Casting a billion nets
One of the first people to begin exploring how evolution creates was the Austrian scientist Peter Schuster. (He is one of those researchers whom one has to label simply “scientist” because it is impossible to decide if he is a physicist, chemist, or biologist.) Because of his interest in the possibility that life began with RNA—in the so-called RNA world—in the 1970s Schuster began to explore what RNAs could do. To act as a catalyst, an RNA molecule needs to fold into the right shape. As with a protein, this shape is determined by the molecule’s sequence of building blocks. In effect this is a genotype-to-phenotype question, with the genotype being the RNA’s sequence and the phenotype its folded shape. The phenotype determines how equipped an organism is for its environment, and it is where selection acts. That selection is ultimately registered in the genotype—the genes that allow a trait to be passed to offspring—but the connection between phenotype and genotype retains many mysteries. Arguably it is the biggest mystery of the post-genomic era we are now entering, in which it is proving unexpectedly hard to identify where many inherited phenotypic traits are encoded in the genotype.
In the 1990s Schuster and his colleagues devised a computer program that could predict the simplest features of an RNA’s shape (its secondary structure, which is how parts of the chain stick to other parts by pairing-up of bases) from its sequence. For an RNA of 100 bases, there are around 1023 possible shapes of this sort. But what was remarkable was the way these shapes and their sequences were related.
Naively, you might expect RNAs with a similar shape, and thus presumably phenotype, to share a similar sequence, so that a map of the possible sequences—the sequence space, which can be represented as a many-dimensional space where each grid point corresponds to a particular sequence—is divided up into various “shape kingdoms” (See Not a Patch, a). But that wasn’t what Schuster found. Instead, RNAs with the same shape could vary very widely in sequence: You could get the same shape, and therefore potentially the same kind of catalytic function, from very different sequences. But these phenotypic siblings weren’t totally isolated from one another in distant parts of the sequence space. Instead, RNAs with the same kind of shape were connected in a network that weaved its way throughout pretty much the whole of sequence space. You could go from one sequence to another with the same shape (and thus much the same function) via a succession of small changes to the sequence, as if proceeding through a rail network station by station. Such changes are called neutral mutations, because they are neither adaptively beneficial nor detrimental. (In fact even if mutations are not strictly neutral but slightly decrease fitness, as many do, they can persist for a long time in a population as if they were quasi-neutral.)
And this was the case for all the 1023 possible shapes. In other words, this RNA sequence space is threaded with that number of interwoven networks, all interlocking but rarely intersecting. This means that if we consider any given sequence at a particular point in the high-dimensional space, it has a huge number of neighboring sequences that have completely different shapes—and yet it can still mutate step by step into a very different sequence with a similar function, provided that it stays on the right track (See Not a Patch, b).
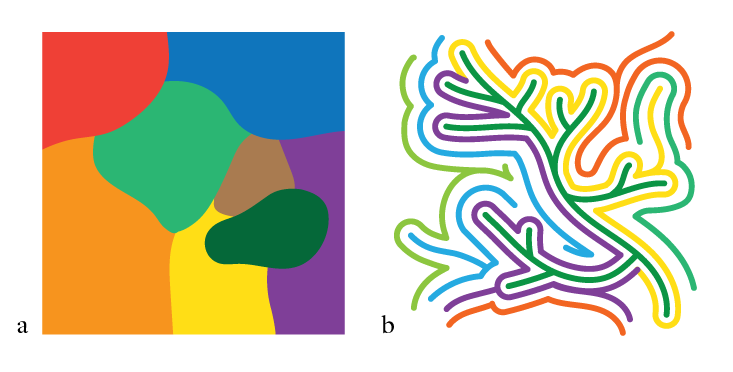
This tells us two crucial things about the RNA sequence space. First, there are many, many possible sequences that will all serve the same function. If evolution is “searching” for that function by natural selection, it has an awful lot of viable solutions to choose from. Second, the space, while unthinkably vast and multi-dimensional, is navigable: You can change the genotype neutrally, without losing the all-important phenotype. So this is why the RNAs are evolvable at all: not because evolution has the time to sift through the impossibly large number of variations to find the ones that work, but because there are so many that do work, and they’re connected to one another.
Susanna Manrubia of the Spanish National Center for Biotechnology has conducted more sophisticated computational studies of how RNA sequence dictates secondary structure, which support this view that there is a lot of redundancy in the genotype-phenotype map.1 She thinks this feature of RNA might have been crucial for its proposed role in the origin of life. “The existence of multiple genotypic solutions, together with a tolerance to exact molecular structures to perform chemical functions, might have been essential in the emergence of an RNA world,” she says.
Proteins have this property too. Different organisms often possess proteins with the same shape and enzymatic function (phenotype), yet typically these will share no more than 20 percent of their amino acids in common. Using a simple model of protein structures, David Lipman and W. John Wilbur of the National Institutes of Health in Maryland showed in 1991 why this should be so: Their simplified model proteins were linked into extended networks in sequence space that can be traversed by neutral mutations one step at a time. In 2001 this apparent redundancy between protein sequence and phenotype was demonstrated experimentally by Anthony Keefe and Jack Szostak at Harvard University. They set out to look for proteins made from randomly assembled amino acids that could bind to the small molecule called ATP, which is the key energy-storage molecule of living cells. Just about any protein that does useful work, such as transporting other molecules or catalyzing their transformation into new forms, uses ATP as the energy source. ATP-binders are therefore vital components of cells. Szostak and Keefe used a chemical method to assemble 80-amino-acid proteins at random from their component parts, and were able to sift through all the variants that they made to find ones that happened to be aptly shaped to bind ATP.2 They couldn’t make anything like the total number of possible permutations of this number of amino acids. But among the tiny fraction (6 trillion) that they did produce, they found four ATP binders. That doesn’t sound like a lot, but if there are already four solutions in this incredibly small sampling of the sequence space, there must be an immense number in the rest of the space—about 1093 of them, the researchers estimated.
This suggests that evolvability, and the corollary of creativity or innovability, is a fundamental feature of complex networks like those found in biology.
Wagner and his coworkers have discovered that this “evolvable” (they call it a robust) structure is a common feature of biological complexity. In 2006 he and his postdoc João Rodrigues began to explore the versatility of the metabolic network of the bacterium Escherichia coli (E. coli).3 This is perhaps the most studied of all bacteria, and the biochemical pathways of its metabolism are well mapped out. E. coli consumes glucose and makes from it all 60 or so of the key molecular building blocks it needs to survive. But what if just one of those metabolic reactions was altered? Could these “neighbors” of E. coli still survive on glucose alone? There is a huge number of such neighbors, but Wagner and Rodrigues calculated the metabolic reactions for more than 1,000 of them, and found that several hundred will also make do with glucose alone. In other words, E. coli’s metabolic network is not finely tuned to run on glucose—lots of other variants will work too. The researchers continued to search through the metabolic space, asking whether neighbors of neighbors would also run on glucose. They found that they could just keep on walking through this network of viable glucose-metabolizers until they reached hypothetical organisms that shared only 20 percent of the reactions with E. coli. Again, the options are extremely numerous, and all reachable by small neutral steps through the space of possibilities. Evolution sends vast numbers of organisms off to explore this intricately ordered library, seeking good answers to life’s problems. As they stumbled upon this unexpected topology of “metabolic space,” says Wagner, “I was close to ecstatic.”
And gene circuits? Same story. Wagner has collaborated with physicist Olivier Martin to explore the space of hypothetical gene regulatory networks. They searched for different networks that, by mutually regulating the activity of the constituent genes, would produce the same overall patterns of gene expression— that is, the same phenotype.4 To take a trivial example, in a network of three genes A, B, and C, the same result of switching off expression of gene B could be achieved by either A or C inhibiting B. Even for just a dozen or so genes the number of possible networks of interactions becomes too vast for an exhaustive search, and so the researchers could examine only a tiny region of the space. But the outcome was clear: On the one hand, circuits typically have dozens to hundreds of neighbors with the same phenotype, while on the other hand circuits that differed in more than 90 percent of their “wires” could still produce the same expression pattern.
This helps to explain a puzzling aspect of gene circuits: their robustness. In the late 1990s a team of researchers at Stanford University created around 6,000 mutants of brewer’s yeast, each of them lacking a different single gene, and found that many of them thrived just as well as the unmutated yeast did.5 The same proves to be true for many other organisms: You can obliterate many of their individual genes to no obvious effect. But this is no surprise if there are plenty of similar gene circuits that do much the same job as the original one. Looked at this way, robustness is complementary to innovation: Any network that can evolve new features and forms among a vast array of alternatives must necessarily be robust against small changes, because it almost certainly has an alternative on hand that performs equally well. This realization offers an antidote to an excessively deterministic view of genes: Exactly which genes you have may not matter so much (within reason), because the job they do is more a property of the network in which they are embedded.
Wagner’s work is “really innovative—no pun intended—and important stuff,” says Johannes Jaeger of the Center for Genomic Regulation in Barcelona, Spain. He is trying to test these ideas by studying the evolution and developmental biology of gene networks that control the growth of insects such as fruit flies. “So far, our results agree with Andreas’ insights,” Jaeger says. They have found that manipulating this network to switch between sequential and simultaneous formation of the segments of a fly’s body is much easier to achieve than expected—it just needs a few tweaks of the regulatory mechanisms, not a wholesale reorganization of genes. “This explains much of the differences in expression patterns in different fly species,” says Jaeger. “All kinds of patterns can be produced by the same set of genes.”
Is evolution a law of nature?
These findings uncover a property of biological systems even deeper than the evolutionary processes that shape them. They reveal the landscape on which that shaping took place, and they show that it was only possible at all because the landscape has a very specific topology, in which functionally similar combinations of the component parts—genes, metabolites, protein or nucleic-acid sequences—are connected into vast webs that stretch throughout the whole of the multidimensional space, each intricately woven amidst countless others.
One might argue that the original creative act of the living world was the generation of the components themselves: the chemical ingredients, such as amino acids and sugars, that comprise the molecules of life. But this now seems like the easy part, the kind of happy accident that chemistry can supply given the right raw materials and environment. The harder question is how one can get beyond that passive soup to kick-start Darwinian evolution. Manrubia thinks that this primal creative step might itself be a consequence of the richness and intimate interweaving of neutral (or quasi-neutral) networks. This means that, even for random, abiotically generated RNA sequences, there is a significant chance of finding ones that perform some useful function. “In a sense, you have function for free if the phenotypes are sufficiently represented in sequence space,” she says. And her computer simulations show that such RNA sequences aren’t rare. “So sufficiently good solutions to act as seeds of the evolutionary process might arise in the absence of the evolutionary process itself.” In particular, there’s a fair chance of hitting on sequences that can replicate—and then you’re up and running. “Natural selection can very quickly turn mediocre solutions into fully adaptive ones,” Manrubia says.
These ideas suggest that evolvability and openness to innovation are features not just of life but of information itself.
The structure of these combinatorial landscapes of biomolecules then enables nature to make bold and creative innovations rather than being forever consigned to making incremental variations on what already exists. Evolution need only take a random walk along a web of neutral (or at least almost neutral) mutations, that, without impairing the fitness of an organism, surrounds it with very different neighbors: innovative solutions to selective pressures that are there for the taking when the circumstances compel it. Through this neutral drift, organisms can reach locations in phase space which would not have been accessible by strictly adaptive mutation from their original starting position.
The role of such random drift in evolution has been long recognized, and recently evolutionary biologist John Tyler Bonner has argued that it may be more important than previously thought, especially for small organisms whose wide variety of shapes and structures—think of the microscopic marine organisms such as diatoms and radiolarians—do not necessarily reflect any adaptive fine-tuning.6 The apparent creativity and artistry of such forms has astonished biologists and inspired artists. Now it seems we can understand where this diversity and inventiveness comes from.
What is more, the new perspective offered by the work of Wagner and others might resolve long-standing disputes about the apparent conflict between the fitness of populations and individuals. Some bacteria seem to undergo more mutations than is “wise” for the individual, if most mutations decrease fitness. An overly simplistic explanation is that many mutations are nonetheless good for the population as a whole because they offer more options for adapting to new environmental challenges. But mutations on robust networks have more chance of being neutral—a feature that is good both for the individual (because it has less chance of incurring deleterious mutations) and for the population (because it provides new ways to adapt when the need arises).7
Yet the question that remains is: Why does the space of evolutionary options have this essential, robust structure? “We simply don’t know why genotype networks are interwoven the way they are,” admits Wagner. Jesse Bloom of the Fred Hutchinson Cancer Research Center in Seattle, a specialist on protein evolution, suggests that perhaps that question is back to front: “One could posit that evolution is only able to work effectively if this property exists, and so the things that ended up evolving have this property,” he says. But he admits that this would be hard to prove.
However, it seems likely that the answer lies beyond biology. Karthik Raman, a former postdoc in Wagner’s lab, now at the Indian Institute of Technology Madras, has studied much the same issues of functional equivalence of different circuits not for genes but for electronic components that carry out binary logic functions. By randomly rewiring circuits of 16 components and figuring out which of them will perform particular logic operations, Raman found that they too have this evolvable topology.8 But crucially, this property appeared only if the circuits were complex enough—if they had too few components, small changes destroyed their function. “The more complex they are, the more rewiring they tolerate,” says Wagner. Not only does this open up possibilities for electronic circuit design using Darwinian principles, but it suggests that evolvability, and the corollary of creativity or innovability, is a fundamental feature of complex networks like those found in biology.
Manrubia agrees that complexity is the key. “It seems clear that efficient navigability can only be achieved in genotype spaces of high dimensionality,” she says. That simply puts more options in reach—because you have more directions to reach in. “As the number of possible neighbors of a sequence increases, the likelihood that some of those neighbors has a viability comparable to the original one grows.” One consequence, she says, is that there could be an adaptive imperative favoring larger genomes, at least for organisms inhabiting varying environments: That way, you gain more in robustness than you lose in the labor of replicating and maintaining a lot of DNA.
These ideas suggest that evolvability and openness to innovation are features not just of life but of information itself. That is a view long championed by Schuster’s sometime collaborator, Nobel laureate chemist Manfred Eigen, who insists that Darwinian evolution is not merely the organizing principle of biology but a “law of physics,” an inevitable result of how information is organized in complex systems. And if that’s right, it would seem that the appearance of life was not a fantastic fluke but almost a mathematical inevitability.
Philip Ball is the author of Invisible: The Dangerous Allure of the Unseen and many books on science and art.
References
1. Aguirre, J., Buldú, J.M., Stich, M., & Manrubia, S.C. Topological structure of the space of phenotypes: The case of RNA neutral networks. PLoS One 6, e26324 (2011).
2. Keefe, A.D. & Szostak, J.W. Functional problems from a random-sequence library. Nature 410, 715-718 (2001).
3. Rodriguez, J.F.M. & Wagner, A. Evolutionary plasticity and innovations in complex metabolic reaction networks. PLoS Computational Biology 5, e1000613 (2009).
4. Ciliberti, S., Martin, O.C., & Wagner, A. Robustness can evolve gradually in complex regulatory gene networks with varying topology. PLoS Computational Biology 3, e15 (2007).
5. Winzeler, E.A., et al. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science 285, 901-906 (1999).
6. Bonner, J.T. Randomness in Evolution Princeton University Press (2013).
7. Wagner, A. The role of robustness in phenotypic adaptation and innovation. Proceedings of the Royal Society B 279, 1249-1258 (2012).
8. Raman, K. & Wagner, A. The evolvability of programmable hardware. Journal of the Royal Society Interface 8, 269-281 (2011).