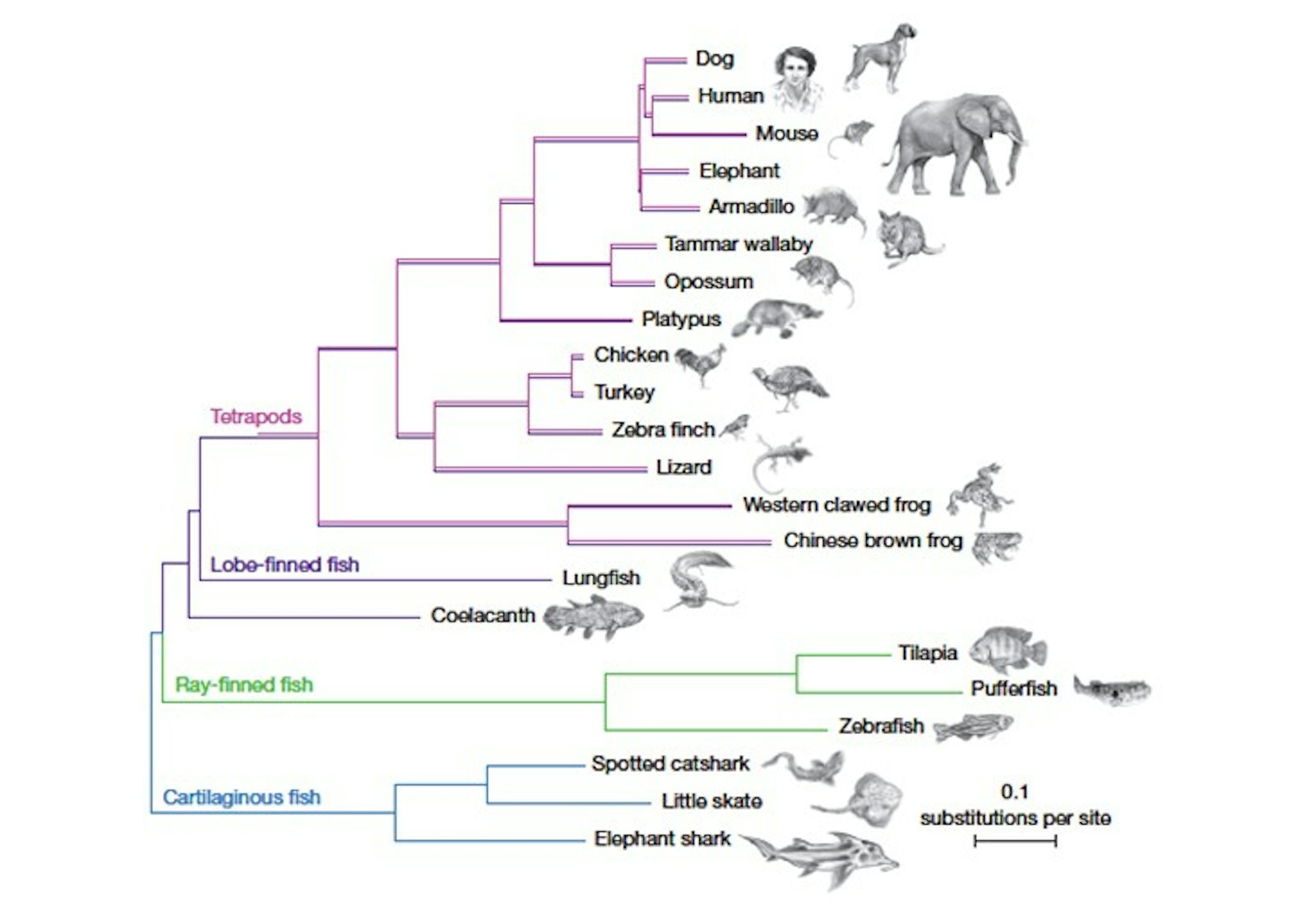
When an enormous four-finned fish surfaced in a South African fisherman’s catch in 1938, scientists were fascinated by its resemblance to fossilized creatures that had died out millions of years ago. The fish, called a coelacanth, turned out to be the first descendant of those organisms ever spotted by humans. The two living species identified since then are so rare, and so elusive, that only a few specimens have ever made their way into labs. But those scant samples have been enough to hook scientists: On the family tree of life, we are more closely related to the coelacanth than to almost all other fish, which means that its genome can provide crucial information about what changed in our ancestors so they could survive on the shore.
The coelacanth’s genome has been hard to get at, as the only tissue scientists have to work with is a few rotting lumps of fish meat, many decades-old, shuttled between freezers and among researchers like jewels. When flesh rots, so does DNA, and most genetics studies on coelacanths have looked only at fragments. But some years ago, Chris Amemiya, a coelacanth researcher at the Benaroya Research Institute in Seattle, realized that a particular sample was just a bit fresher than others, and could provide the basis for the first attempt to sequence the full genome of a coelacanth. A team of nearly 100 scientists, including Amemiya and led by a group at the Broad Institute in Cambridge, Mass., undertook the task, and their paper on the genome of Latimeria chalumnae, the African coelacanth, appeared in Nature last month.
Several things about the coelacanth genome jumped out at the researchers. The analysis made it clear, for instance, that the coelacanth is less closely related to the first land-dwelling organisms than is the lungfish, settling a long-running question. (The lungfish’s enormous genome poses great obstacles to sequencing, however, so for now the coelacanth is our closest fishy relative with a sequenced genome.) And certain parts of the coelacanth genome seem to evolving unusually slowly, which interests biologists.
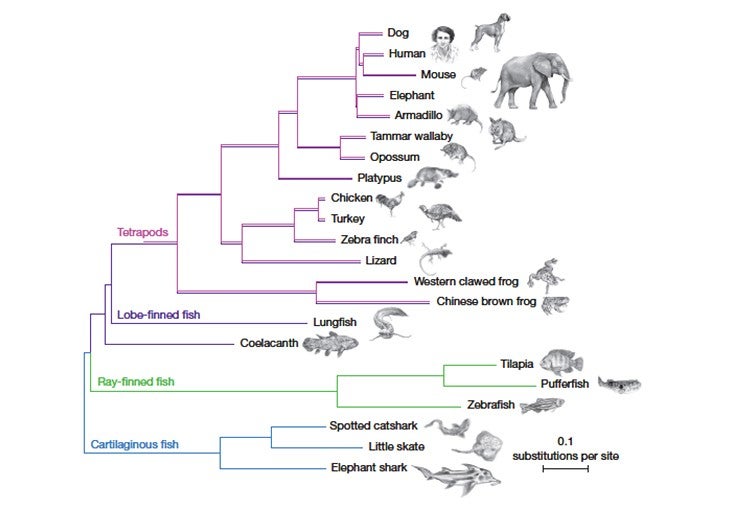
The researchers also identified certain genes shared by the coelacanth and all bony fish but not present in land vertebrates, which means that the genes must have been lost after our ancestors diverged from the coelacanth’s ancestors. In addition, they pinpointed several categories of genes and gene regulators, including those related to limb development, sense of smell, kidney function, and the immune system, that may be related to the transition to land.
In one especially thought-provoking experiment, the researchers took a snippet of coelacanth DNA—a piece whose purpose was unknown but nevertheless showed up in land vertebrate genomes as well—and inserted it into a mouse embryo. There, it switched on a suite of genes that help grow the bones of the ankle, wrist, and digits. The fact that this foreign bit of DNA has that effect in the mouse means that it may do something similar in coelacanths and in land vertebrates like ourselves, the researchers say—though greater certainty will have to wait for later experiments.
It’s easy to forget that there are many systems like this piece of DNA and the genes it regulates—highly evolved, configured to accomplish very specific tasks, yet shared among very different-seeming creatures. Organisms as different as mice and enormous deep sea fish draw from the same set of blocks, genetic LEGO modules that can be used over and over in vastly different combinations. We are all amalgams of earlier creatures, carrying the traces of the past in every cell.
Veronique Greenwood is a former staff writer at DISCOVER Magazine. Her work has appeared in Scientific American, Popular Science, and the sites of Time, The Atlantic, and The New Yorker. Follow her on Twitter here.