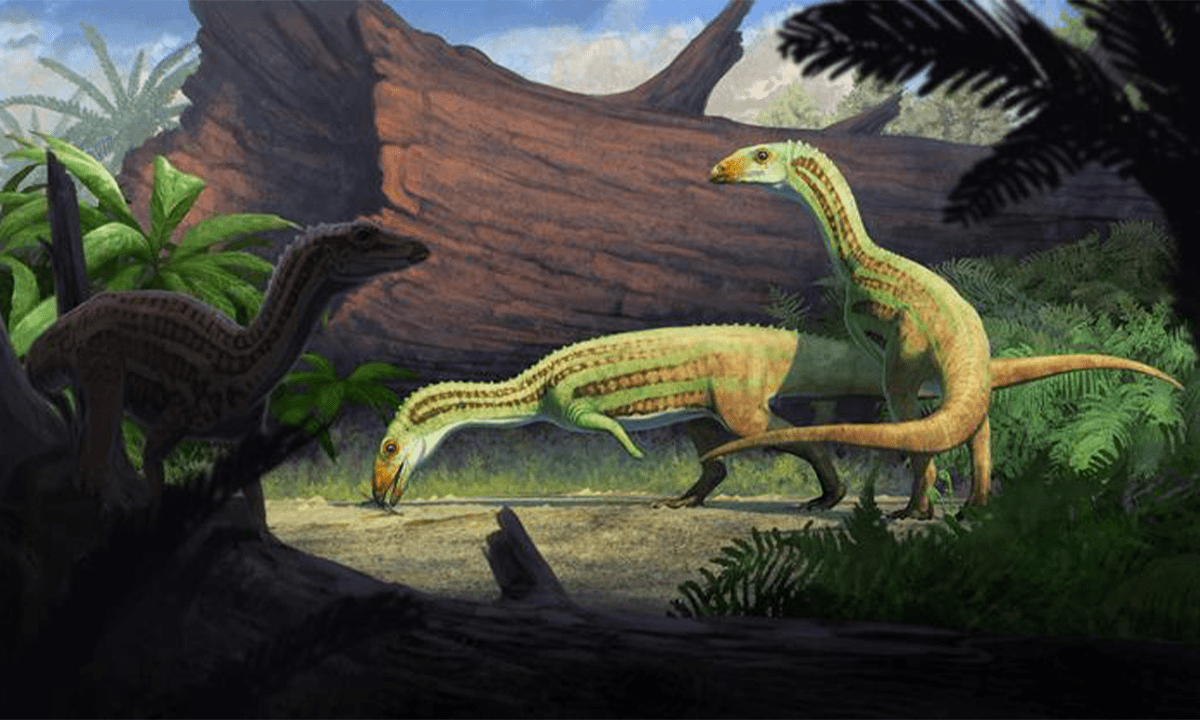Latest
Were You Born to Love Music?
How you respond to art—from poetry, to visual art, to music—may be partly written in your DNA
Latest Stories
Baby Boomers Are a Transition Generation in Our Longevity Crisis
Lifespan in the United States plateaued over a decade ago
Are You Smart Enough to Avoid Falling for “Corporate Bullsh*t”?
New research points to a troubling relationship between buzzwords and decision-making
This Ancient Crocodile Ancestor Learned to Walk on Two Legs
It was a childhood crawler before it walked upright
Psychology
See more PsychologyLaughing Off Your Mistakes Makes You Seem More Competent
“People often overestimate how harshly others judge their minor social mistakes”
Why You’re More Likely to Develop AI-Psychosis than to Join a Cult
Philosopher Lucy Osler on the insidious appeal of AI Chatbots
Blame Your Parents for Your Extreme Aversion to Snakes
But recognize that it’s a useful survival trait
Environment
See more EnvironmentThe Ancient Cold Snaps That May Have Shaped Human Evolution
Here’s when Earth’s climate became chaotic
Is a Strictly Enforced Fishing Ban Saving the Yangtze?
Ecologists detect promising, early signs of river recovery
The Long, Dirty History of Our Capitol’s Waters
The recent Potomac River disaster follows centuries of pollution—but things are looking up
Get unlimited, ad-free Nautilus. Become a member today.
History
See more HistoryHow Three Students Designed an Atomic Bomb
A top-secret 1960s project tasked physics postdocs with building The Bomb
The Thrill of Science in 2042
A science historian explains how science got its groove back. A fictional dispatch from the future.
How Energy Politics Played Out on the White House Roof
The quick removal of Jimmy Carter’s futuristic solar panels echoes more recent feuds over renewables
Physics
See more PhysicsPhysicists Uncover How Long It Takes to Get the Last Drop of Syrup
How to tackle a common kitchen problem with fluid dynamics
Communication
See more CommunicationWhat Would Richard Feynman Make of AI Today?
The scientific sage was always suspicious of grand promises delivered before details were understood
Get the Nautilus newsletter
Cutting-edge science, unraveled by the brightest living thinkers.
Sociology
See more SociologyThe Vibes Have Been Off in the US for Decades
New survey analysis reveals a sense of national deterioration
Does Belief in God, not Political Party, Drive Conservatism?
Religious “nones” may be less socially liberal than they used to be
Parachute Science Continues to Prevail in Global South Biodiversity Studies
The privilege of describing new species is skewed to Global Northerners
Some Memories Live in the Brain Even If We Can’t Recall Them
It’s all coming back to me now
More recent posts
See all postsRestoring Panama to When Prehistoric Beasts Roamed the Jungle
The Central American nation never fully recovered from the loss of its megafauna
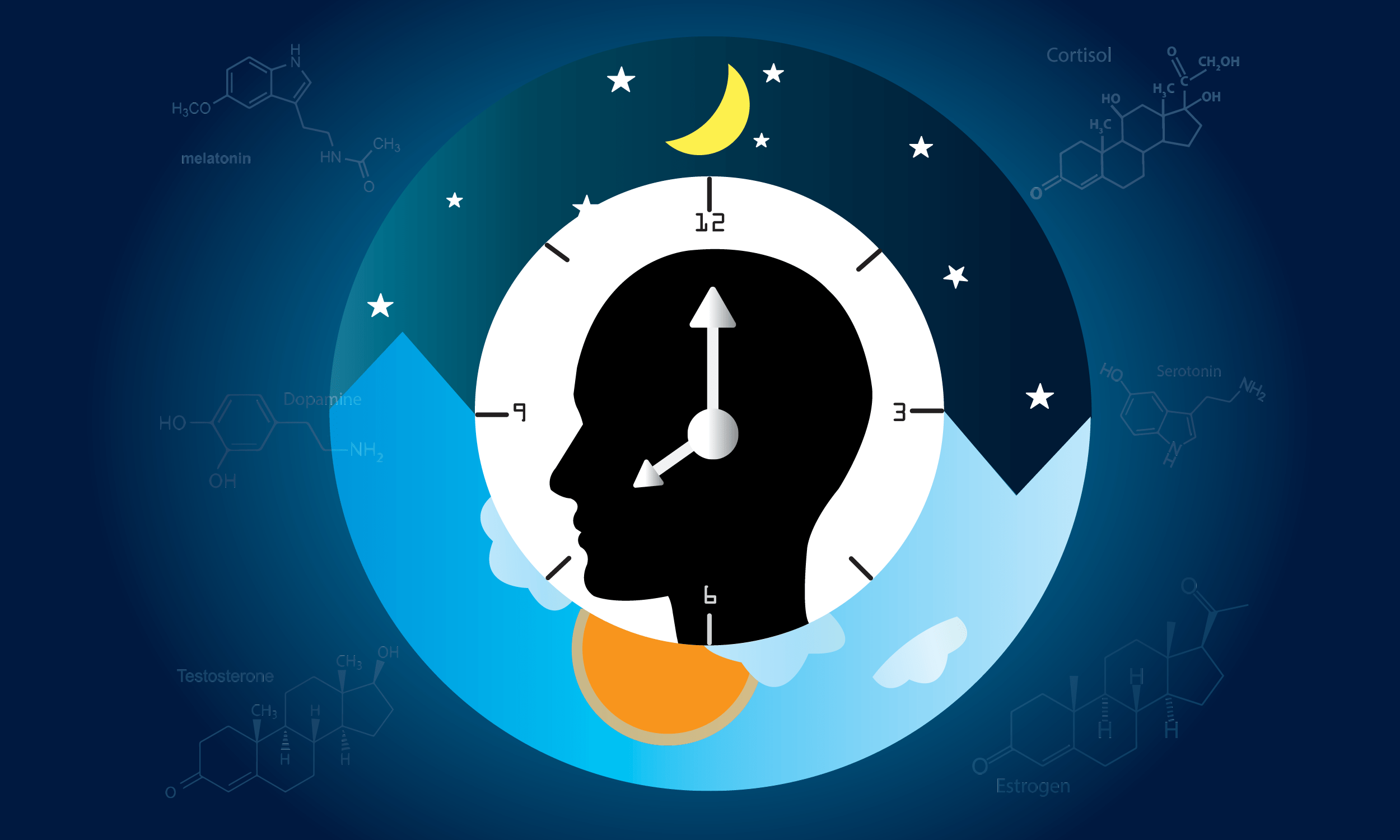
Your Biological Clock is More Complex Than You Think
Prepare for Daylight Saving Time by taking a tour of your internal timekeeping machinery
The Dainty Dinosaur That’s Rewriting Evolutionary History
New alvarezsaur fossil offers a Cretaceous missing link
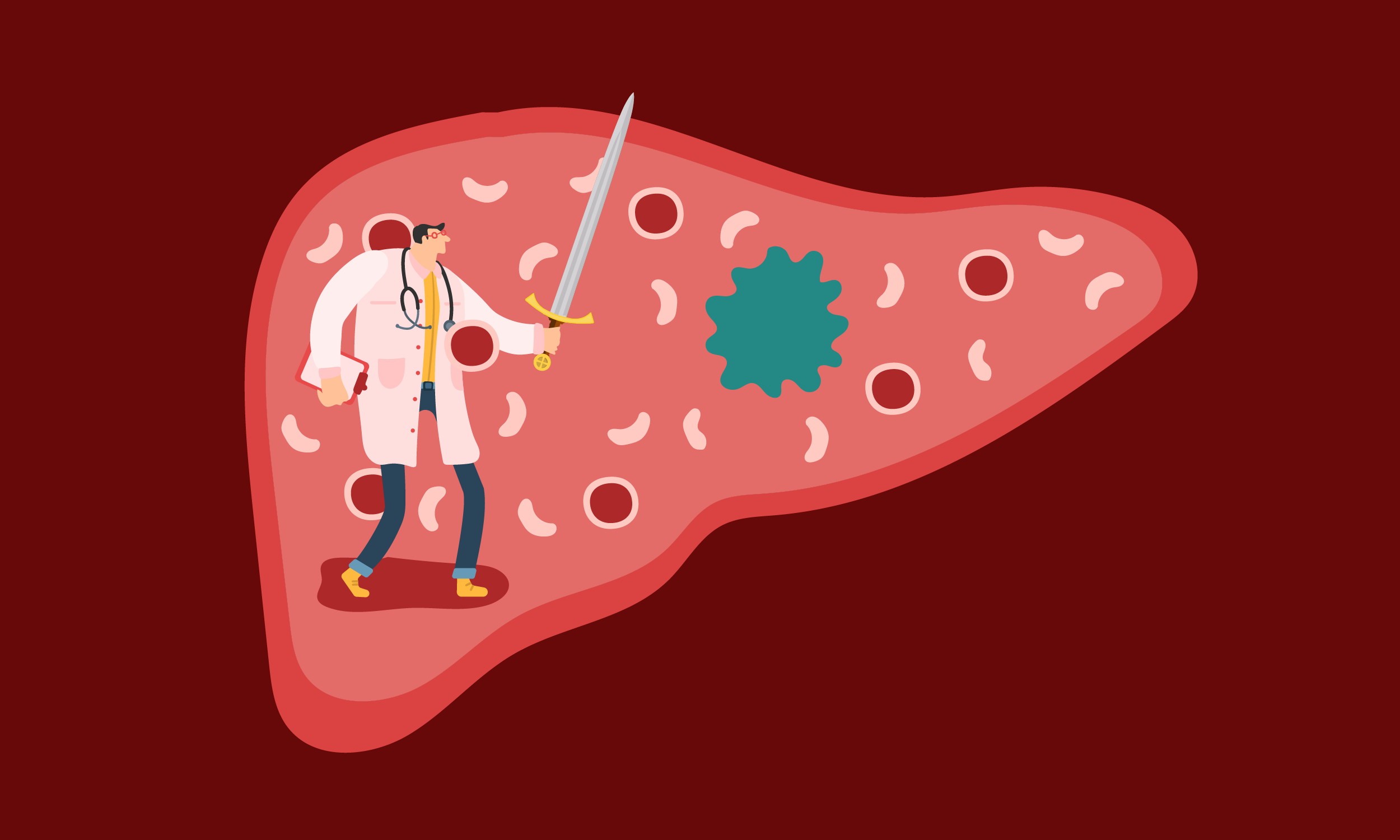
These Bacteria Beat Cancer By Eating Cancer
They’re being engineered to devour tumors from the inside out
Koalas Recover Genetic Diversity as Populations Expand
Their rebound shows resilience after a severe genetic bottleneck
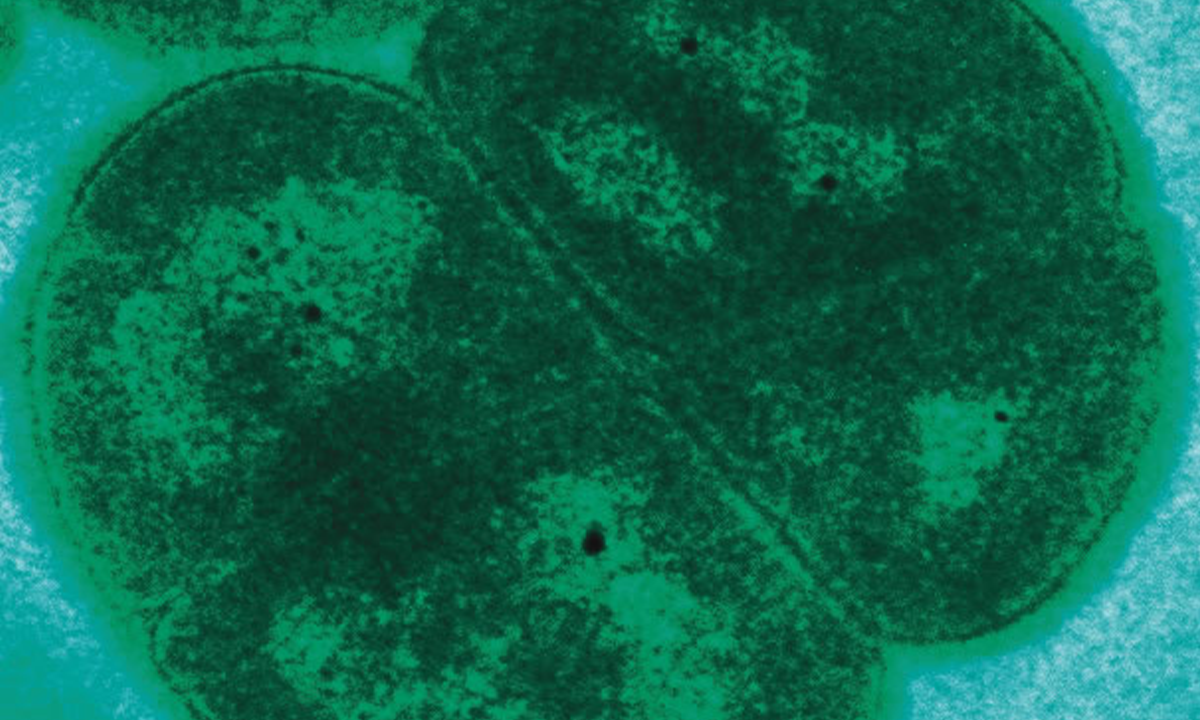
Watch How Planet-Hopping Microbes Can Survive Asteroid Strikes
This polyextremophile bacterium could survive an interplanetary trip
When Scientists Are Dinosaurs
At the paleontology conference, her new theory was shouted down
Paleontologists Solve the Mystery of a Twisted Jawbone With Sideways Teeth
The Tanyka was a little like a platypus