Why don’t we have an arsenal of fast-acting cures for tuberculosis, malaria, and pneumonia? In part it’s because scientists can’t fully understand what they can’t observe: Namely, the way the pathogens that cause diseases and infections live within the human body. The real homes of pathogens—in the blood and tissues of a host organism—are impossible to replicate, so researchers learn what they can by studying these microscopic organisms in petri dishes and test tubes.
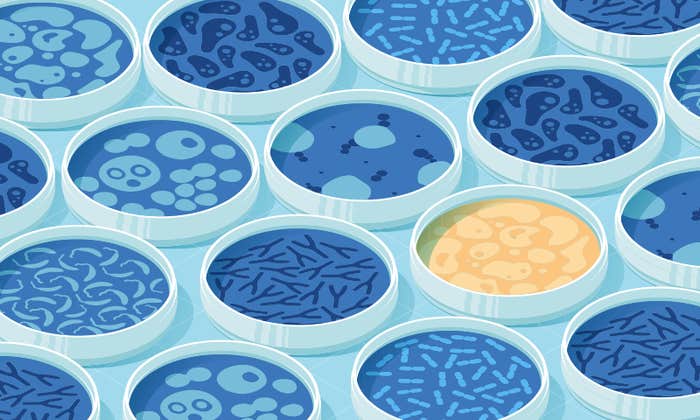
These artificial homes for bacteria have been around since the late 19th century, when biologist Robert Koch grew colonies of the bacteria that causes tuberculosis, Mycobacterium tuberculosis, in glass test tubes. These vials, and later, glass plates, filled with a gelatinous mix of agar and nutrients, soon filled microbiology labs. “It allowed scientists to identify what bacteria were causing awful, awful diseases like tuberculosis because you could isolate colonies,” says Jason Shear, a chemist at The University of Texas, Austin, “but for the most part the technologies haven’t dramatically advanced.” The technical term for this kind of experimental environment is in vitro, a Latin phrase that translates as “in glass.”
Much of what microbiologists know about bacteria comes from in vitro experiments, but sometimes the results say more about how pathogens live in the lab than in people. The pharmaceutical company Novartis learned this lesson the hard way in 2009, when they invested heavily in a drug that annihilated M. tuberculosis in the lab before they found that it could not save mice with the disease. One year later, they realized the drug killed M. tuberculosis by turning glycerol in the test tubes toxic, but because glycerol is not abundant inside bodies, the drug had no effect on the pathogen in real infections. Just as a person’s vulnerability to attack changes depending on where they live—in a fortress surrounded by hundreds of other people or camped alone on a hill—a pathogen’s vulnerability to attack depends on where it makes its home.
In reality, M. tuberculosis constructs its abode inside cells. Shortly after a person breathes the microbe in, it infects one of their immune cells, where it can wait, protected by a waxy coating, for years, even decades, until the time is ripe for replication. When its victim’s immune system weakens, the microbe multiplies and its progeny burst out of the immune cell, infecting other cells. As the infected cells die, M. tuberculosis builds a fortress out of greyish cell carcasses, forming lumps in a person’s lung and causing them to gasp for air.
Scientists can’t fully understand what they can’t observe: Namely, the way the pathogens that cause diseases and infections live within the human body.
Researchers studying M. tuberculosis in vitro have not found drugs that can efficiently penetrate the lumps the microbe builds in human lungs, because these structures can’t be replicated in the lab. “So much of the way we culture bacteria in vitro is completely artificial,” says Clifton Barry, chief of the Tuberculosis Research Section at the National Institute of Allergy and Infectious Diseases in Bethesda, Md. “It has nothing to do with what it looks like in humans.” As a result, the most effective treatment for tuberculosis is a grueling six-month course of chemotherapy.
Bacterial homes can be defined in multiple ways. In addition to the physical barriers they create within living tissues, homes can also be built out of bacterial communities. Matthew Parsek, a microbiologist at the University of Washington, studies the ways in which some bacteria rely on their neighbors to fight medicines and the human immune system.1 One common member of these communities is Pseudomonas aeruginosa. It often lodges itself in layers of mucus within the lungs of people with cystic fibrosis—a genetic condition that prevents cilia in the lungs from removing infectious microbes. P. aeruginosa manufactures chains of sugar molecules that help it cling to nearby bacteria, such as Staphylococcus aureus. Certain strains of S. aureus, lumped together under the acronym MRSA, resist several antibiotics and kill roughly 11,000 people in the United States each year. Together, P. aeruginosa and S. aureus are extremely difficult to kill because they combine their individual abilities to fend off antibiotics.

Parsek likens the arrangement between the microbes to living in an apartment. “If you have one roommate that’ll do a certain job and you do another job, it might be a pretty darn favorable arrangement,” he says. Inside the tissues they infect, P. aeruginosa and S. aureus form small, dense colonies of only a few hundred or thousand microbes. In infected wounds, colonies are spaced regularly and spread deep into the tissue. The bacteria “talk” with one another through chemical messages, but their ability to communicate is affected by their proximity to one another and by the porousness of the tissues they infect. However, the details of their interactions are poorly understood because researchers cannot control how the bacteria grow in petri dishes and test tubes. Scientists are only beginning to understand how chemical messages between bacteria work, and Parsek suspects that the ways bacteria arrange themselves in three dimensions affects the way they communicate and defend themselves.
Certain strains of S. aureus, lumped together under the acronym MRSA, resist several antibiotics and kill roughly 11,000 people in the United States each year.
The intricacies of the bacterial home have resisted the reach of science for more than a century. But this may finally be changing. In October, Shear and co-investigator Marvin Whiteley, a microbiologist at The University of Texas, Austin, reported on a new way to make more natural abodes for bacterial communities. They took a cue from 3D printing, which is an additive fabrication process that pieces together a series of 2D cross-sections to build an object of arbitrary shape. Shear and Whiteley used a related subtractive process that began with mixing bacteria and a laser-absorbing dye into the main ingredient in Jell-O brand desserts: gelatin. The mixture hardens when struck by a laser beam. By moving a beam through the material, the team sculpted three-dimensional chambers that hold volumes smaller than a picoliter, or one millionth of a millionth of a liter. These chambers provide precisely measured spaces for colonies of as few as a hundred or so individual bacteria, allowing bacterial density to be measured and controlled. In the October report that debuted their 3D homes, Shear and Whiteley demonstrated that tightly packed colonies of P. aeruginosa are more resistant to antibiotics than looser groupings.2
The gelatin-walled chambers have another advantage: They are chemically permeable, which allows bacteria to communicate with nearby communities. While the bacteria remain trapped inside their chambers, nutrients and chemical messages from bacteria in other chambers can filter through the hardened gelatin as they do through infected tissues. This allows experimenters to arrange the chambers in three-dimensional patterns like the bacterial colonies in an infection and test new ideas about how bacteria communicate and resist antibiotics.
Researchers look forward to seeing what Shear and Whiteley find with their 3D creations. “They’ve been able to develop this new system that allows them to ask questions that most microbiologists can’t,” Parsek says. Whiteley plans to use this technology to recreate the way that bacteria arrange themselves within diabetic patients’ wounds, which often become badly infected. Other researchers may sculpt the gelatin into whatever shape mimics their favorite pathogen’s home in a person’s lungs, skin, or any other organ. Barry believes that the technique could be beneficial to tuberculosis research. “A lot of treating the disease involves many assumptions about what the bacteria experience at the time that you are treating them,” he says. “We could benefit from looking at bacteria in a more confined space and under more physiologic conditions.”
Whiteley and Shear may have engineered a new petri dish for the 21st century. And in doing so, they have affirmed for bacteria what we already know for ourselves: home matters.
Zach Zorich is a freelance science journalist and contributing editor at Archaeology magazine.
References
1. Hibbing, M.E., Fuqua, C., Parsek, M.R., Peterson, S.B. Bacterial competition: Surviving and thriving in the microbial jungle. Nature Reviews Microbiology. 8(1), 15-25 (2010).
2. Connell, J.L., Ritschdorff, E.T., Whiteley, M., & Shear, J.B. 3D Printing of microscopic bacterial communities. Proc. Natl. Acad. Sci. U.S.A. 110, 18,380-18,385 (2013).