Nir Hacohen, an immunologist and geneticist at the Broad Institute of the Massachusetts Institute of Technology and Harvard University, knew that biology had a problem. He wanted to understand the human immune response’s role in cancer and other diseases. But to do that, he first had to address a more fundamental issue: The definition of the immune cell types themselves seemed insufficient, incomplete, and outdated.
For over a century, distinctions between types of cells relied on how they appeared under a microscope: their shapes, sizes, locations, and their uptake of staining dyes. Recent decades, however, witnessed a shift to molecular methods that use fluorescently labeled antibodies to target protein markers on the cell’s surface. Although this approach allowed researchers to isolate more cell types, it was not enough, according to Hacohen. Until 2009, biologists could analyze cells only in bulk, averaging signals from multitudes of them to get a picture of what was going on in a tissue. When sequencing RNA from individual cells finally became possible, the initial analyses were what Hacohen called “biased” and “shallow” because the few markers used to classify the cells were too insensitive to nuances of differences among them. “Does this really capture the complexity of the cell?” Hacohen said.
In a study published in Science this past April, he and his team showed that, as expected, much of this complexity had been obscured. Analyzing patterns of gene expression in individual human immune system cells, the researchers refined the definitions of the types known as dendritic cells and monocytes and identified a novel type that had been overlooked. Moreover, they discovered that a cell population thought to comprise one subtype was actually a mixture of two, which perform different functions.
Hacohen’s work represents one component of a much larger project. Last October, an international community of researchers led by Aviv Regev of the Broad Institute and Sarah Teichmann of the Wellcome Trust Sanger Institute launched the Human Cell Atlas to apply this kind of modeling to the entire body. It aims to catalog not just cell types—which are predicted to extend far beyond the 200 types most often cited in textbooks—but also the hallmarks of cell types under different conditions and in individuals with different genetic and epigenetic variations. That knowledge is important because it would provide a more comprehensive overview of the dynamic complexity of life. Immune cell subtypes might shift in someone who has an infection or an allergy or an autoimmune disease, for example; or they may vary for different population groups.
“This is not comparable to the Human Genome Project,” Hacohen said. “That was a fairly well-prescribed problem. Here the problem is much more difficult and in a sense encompasses a lot of biology.”
The Human Cell Atlas is only one of several projects in molecular and cellular biology looking to synthesize enormous quantities of data to gain deeper insights into just how diverse the cells in our bodies really are, and how complex life is. In 2003, researchers at the KTH Royal Institute of Technology, in Sweden, launched the Human Protein Atlas, which aims to catalog comprehensively the expression, location, and spatial distribution of proteins within individual cells. Only within the past few years were members of the project able to start classifying, annotating, and analyzing the millions of images they had captured of subcellular structures in different cell types. To reach that point, they first had to spend a decade standardizing, optimizing, and scaling up their procedures, which involved using targeted antibodies to stain proteins and then looking for those markers inside healthy and cancerous tissue cells with high-resolution microscopy.
In January 2015, the team charted protein expression across more than 30 human tissues. This past May, they published the second part of their undertaking in Science. Turning their attention to the single-cell level, they mapped more than 12,000 proteins to 30 subcellular structures, in turn defining the proteomes—the complete sets of expressed proteins—of more than a dozen major organelles. The researchers identified which proteins were found where, explored variations in protein expression from cell to cell, and analyzed how cells segregate chemical reactions within themselves.
One of the paper’s most salient findings, according to its principal investigator, Emma Lundberg, was that as many as half of our proteins can be found in multiple compartments of a cell. “Everything that proteins do is specific within the context of their environment,” Lundberg said. “If one protein is present in the nucleus but also in the plasma membrane, it might have different functions in those compartments.”
Take HER2, a receptor protein often overexpressed in certain breast cancers. When found in tumor cell membranes, HER2 correlates with a better prognosis than when it is in the cytoplasm or nucleus. “There are more and more and more studies of single proteins showing that this is actually a common phenomenon,” Lundberg said. “But it’s the scale of it,” she added, that is most exciting.
As much as 50 percent of the proteins that her group observed were expressed in more than one part of a cell. If that figure indicates how big multi-functionality could be, Lundberg said, “it makes the cell much more complex and the functionality of the proteome greater.”
This heterogeneity offers deeper insights into the fundamentals of protein function, but it may also explain why, for instance, certain drugs result in unwanted side effects.
Another group of scientists, who hope to publish their work in the fall, have been mapping the distribution of proteins in the cell types of the testis—home to the greatest number of uniquely expressed protein-coding genes. In doing so, they are reclassifying the cell subtypes that occur during spermatogenesis. “Many things are happening in these cells before they become mature,” said Cecilia Lindskog Bergström of Uppsala University in Sweden, who is collaborating on the research. “Proteins that are expressed in a certain sub-stage of sperm development will tell more about the function of these proteins.”
This dynamic way of defining cell type is what Hacohen sought to establish further in his study of blood cells. In the findings it reported in May, the Human Protein Atlas began to demonstrate why these refinements may be necessary. The team observed that approximately 15 percent of the proteins exhibited single-cell variation: In a tissue that looked superficially uniform, some cells might differ from their neighbors in the amount or spatial distribution of the proteins they expressed, when one would expect them to be the same. The single-cell RNA sequencing approach of the Human Cell Atlas will allow researchers to create cell profiles based on molecules other than proteins.
“In the past, we typically looked at a tissue or an organ in the way you’d look at a smoothie,” said Bart Deplancke, a biological systems engineer at the École Polytechnique Fédérale de Lausanne in Switzerland. Based on its overall color and taste, one might assume that a smoothie consists of strawberries and bananas. But that way of looking at it may miss key ingredients and makes it seem as if all parts of the smoothie are identical. With modern techniques, Deplancke said, they can do the tissue-analysis equivalent of looking at a smoothie and saying, “I see these different pieces of fruit.” And they can see how that full diversity of cell types makes a functional organ. Similarly, they can learn how the full spectrum of cells involved in cancers and other diseases relates to prognosis and recovery.
Deplancke is one of three researchers who have begun organizing the Fly Cell Atlas, which seeks to characterize all the cell types in Drosophila fruit flies. The Allen Institute in Seattle is working toward a similar understanding of the mouse brain. Both hope to apply their findings to explain human behavior and disease, just as the Human Cell Atlas does. Ultimately, integrating the vast datasets generated by these different atlases may prove the greatest challenge of all—but, the researchers hope, it will also be the most rewarding, combining structural, genomic, and epigenetic approaches under the umbrella of a new kind of cartographic exploration.
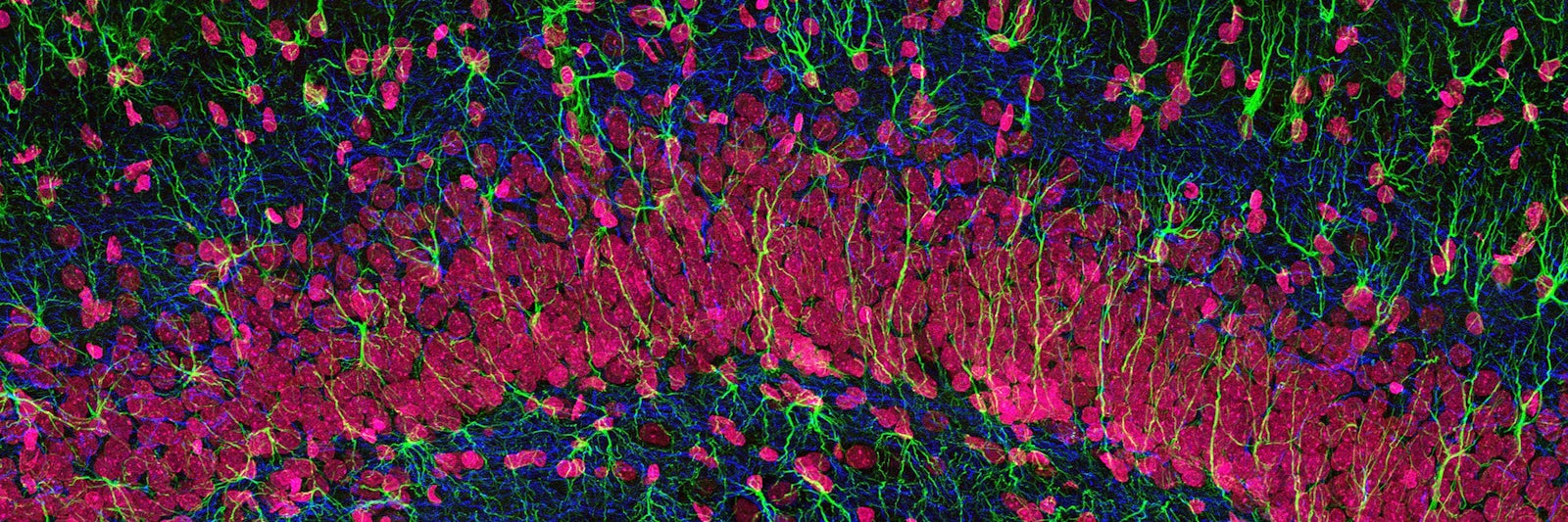
Lead Image: A stained microscopic section hints at the variety of cells in the hippocampus of the brain. But modern analytic techniques have exposed that many tissues hold a vaster diversity of cell types than anticipated—a fact that has important implications for their biological functions. Credit: Thomas Deerinck, NCMIR.