The idea that life’s deepest, oldest roots were laid down by RNA molecules that evolved ever more complexity has dominated the origins-of-life field for the past few decades, reigning over competing theories that started instead with peptides or DNA.
But recently, the field has shifted toward theories that include more than one protagonist. One that’s gained particular momentum is the idea that RNAs and peptides coevolved complexity, and that their intermingling sparked life as we know it.
Now, a study published in Nature breathes fire into an “RNA-peptide world” by suggesting a plausible pathway for how early RNA molecules may have enabled peptides to grow directly on them, like mushrooms growing on a tree. Those peptides may in turn have stabilized the RNA molecules, allowing them space to complexify. This coevolution of two of life’s key players as a single mixed, “chimeric” molecule may have been the very start of protein production, and a step toward a primitive version of a ribosome.
“It turns out that they could have actually helped one another,” said Claudia Bonfio, a junior group leader at the Institute of Supramolecular Science and Engineering in Strasbourg, France, who wrote a commentary that accompanied the paper. Studies have shown that the raw materials for both peptides and RNA would likely have been present at the start of life, so this paper takes the view, “Why should we just focus on RNA?” Bonfio said.
The work opens new directions to explore the origins of life, said Yitzhak Tor, a professor of chemistry and biochemistry at the University of California San Diego, who was not part of the study but is a longtime collaborator with the authors. “Now you need to consider the interplay between two different biomolecules.”
It’s a “highly intriguing demonstration,” said Andro Rios, a research scientist at the Blue Marble Space Institute of Science and NASA’s Ames Research Center.
The findings also explain a major chicken-and-egg conundrum that has bedeviled researchers studying life’s origin: How did proteins form before the evolution of the ribosome—the cellular machine that churns out proteins in modern cells—when the ribosome is partly made up of proteins itself?
Living Fossils
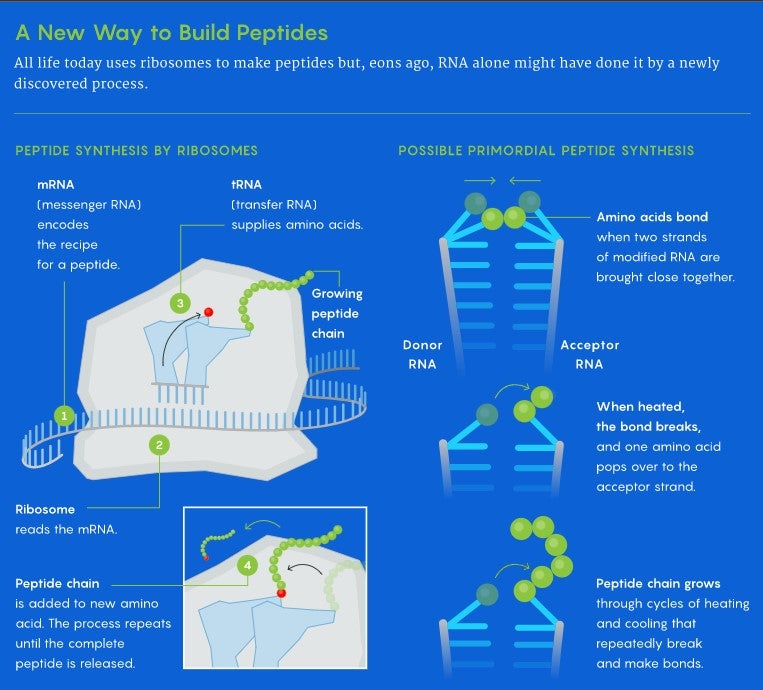
When cells need to make proteins, their genes spin out long threads of messenger RNA (mRNA) that encodes very precise recipes for making them. Ribosomes slide over these recipes to read them and assemble the amino acids that correspond to each step, with the help of their amino acid suppliers: molecules called transfer RNA (tRNA) that continuously dive in as the process unfolds. The ribosome transfers a growing peptide chain onto each new amino acid brought in by tRNAs. These chains get longer and longer and eventually fold into functional proteins.
It’s unlikely, however, that proteins formed in a similar way 3.5 billion years ago. The bonds that hold the peptides on the tRNA molecules are rather weak. Without a ribosome available to provide shelter, water molecules would break these bonds before peptides could form, making the process unfeasible in the harsh watery conditions of the primordial world.
But most people are focusing on re-creating the familiar process of protein translation in a simpler form, said Thomas Carell, senior author of the new paper and the chair for organic chemistry at the Ludwig Maximilian University of Munich. What if ancient translation looked very different from its modern form?
Carell and his team started to dig into this idea, looking for stronger bonds that might have survived in primordial conditions. That’s how they zoned in on the often-overlooked molecules called noncanonical nucleotides. The genetic code of RNA is typically written with just four bases (adenine, guanine, cytosine, and uracil), but nucleotides with other bases are also present in many RNA molecules that do other jobs—including helping with protein production in the ribosome. These unusual nucleotides can attach to amino acids with a chemical bond that’s much stronger than the one that attaches amino acids to the tRNA molecules.
In a 2018 study, Carell and his team reported that the classic nucleotides and the noncanonical nucleotides likely all evolved at the same time before the dawn of life. Some of those in transfer and ribosomal RNAs were present in the last universal common ancestor of all organisms.
Here were “relics of an old RNA world sitting next to each other directly at the oldest part of the system,” Carell said. “So we said, OK, these are our fossils—and let’s see what the fossils can tell us.”
They developed a model for a different kind of peptide-growing process. They imagined two RNA strands capped by these unusual nucleotides, one that they loaded with an amino acid using the stronger bond. After getting the first amino acid to hop to the second strand, they could reload the first strand with another amino acid. By cyclically heating and cooling the system, they could repeatedly break and make bonds between the amino acids on the two chains, flipping the amino acids from one strand to the other and extending the chain.
In effect, if building a peptide is like assembling a tower from building blocks, the new process adds amino acids by adding blocks to the top of a growing stack, while ribosomal protein synthesis extends the tower by moving it atop new pieces at the bottom.
Growing Peptides
Testing their theory involved a set of experiments that Carell called a “tedious” and “brutal tour de force”—but in the end, they showed that the process could indeed produce peptides up to 13 amino acids long.
The process falls far short of the protein translation seen in cells. The most important missing feature is that ribosomes are translating instructions for a specific protein encoded in the mRNA. In the novel system, “we grow relatively random peptides,” Carell said.
But the researchers did succeed in showing that peptides could be built up purely by RNA in a stepwise manner, which is “something that had not been done before,” Bonfio said. All in all, the demonstrated process is a vital step toward molecular recognition, she said, even though it’s not a primitive version of a ribosome.
“So we said, OK, these are our fossils—and let’s see what the fossils can tell us.”
“It’s a really beautiful example of chemistry,” said Sara Walker, an associate professor at Arizona State University who was not part of the study. But she and Lee Cronin, chair of chemistry at the University of Glasgow, both thought that the system might be over-engineered or too unrealistic to mimic what could have happened at the start of life.
For others, however, the work’s mimicry of the primordial world is less important than the doors it opens to further study. Because the findings show that a peptide and an RNA can evolve together, it’s clear that “this can experimentally and chemically be done,” said Nizar Saad, a research assistant professor at Nationwide Children’s Hospital’s Center for Gene Therapy in Ohio. An RNA-peptide world “is what the scientific community is going towards now,” Saad said.
An RNA-Peptide World
“I don’t want to replace the RNA world theory,” Carell said. But “I think we need an extension” to make it more plausible. He thinks that rather than evolving their complexity separately, RNA and peptides did it together as a single molecule, complementing each other’s functions.
A coevolving chimera of RNA and peptides would offer the best scenario for the evolution of life, Carell said. He and his team found not only that the RNA molecules were helping the peptides grow, but that the peptides were bringing stability to the RNA molecules.
As the structure of the chimera eventually got longer and more complex, the peptide portion might have stabilized the RNA enough for it to start self-replicating and evolving. Meanwhile, the RNA might have let the peptide part acquire a structure sophisticated enough to allow it to start catalyzing chemical reactions. Eventually, the parts could have split apart and started to interact in ways that more closely resembled what happens in a ribosome.
Carell and his team are next hoping to learn if they can get their unorthodox RNA molecules to grow specific peptides from encoded information. They then hope to see whether the peptide can develop catalytic functions that might help the RNA to replicate.
Whatever successes or failures lie ahead, it will always be hard to know exactly what happened billions of years ago. “We cannot go back in time, so whatever you construct in the field, somebody can always say, I think it all happened differently,” Carell said. “And if somebody comes up with a better model, [it’s] more than welcome. This is how science develops.”
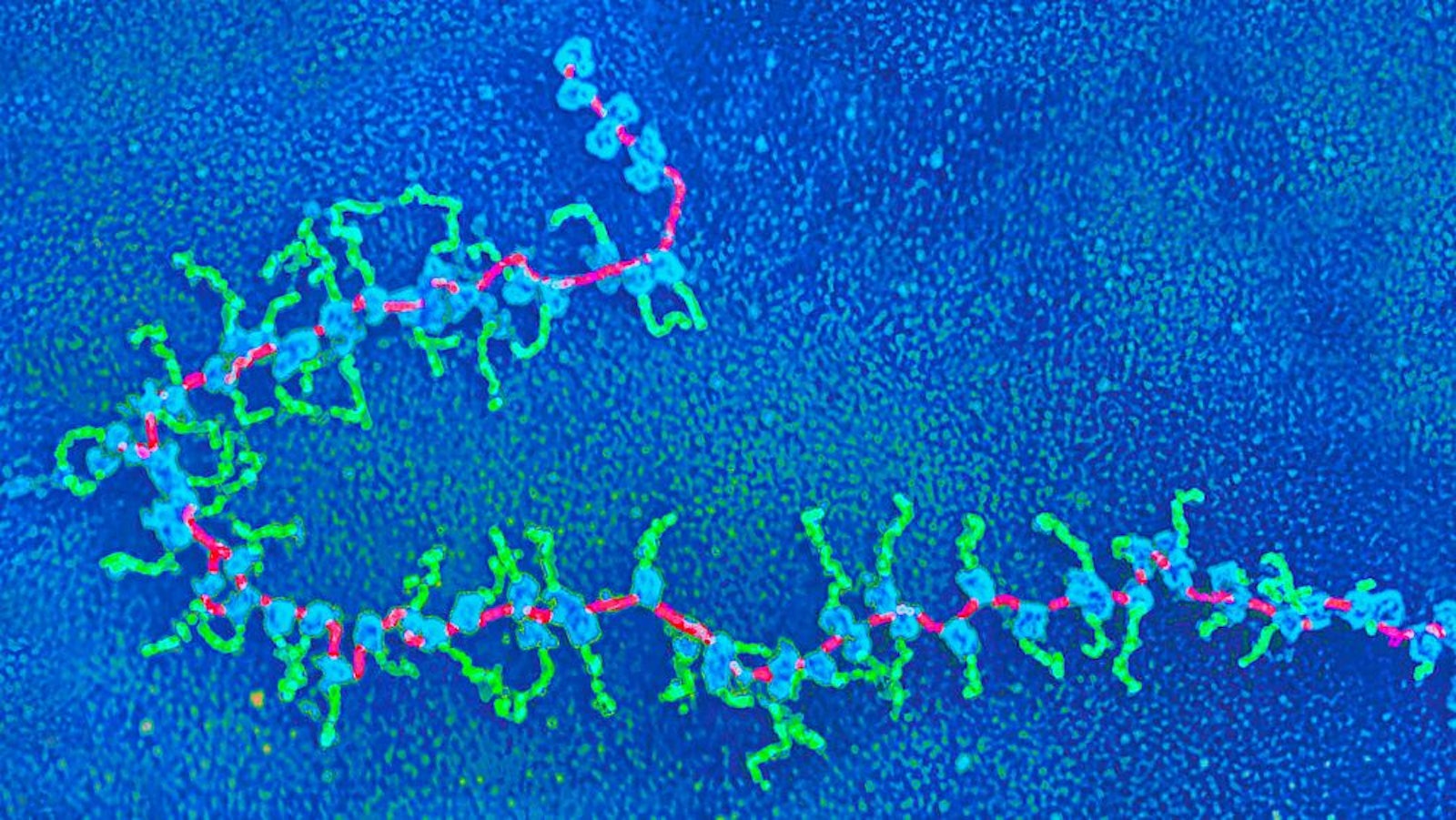
Lead image: Structures in living cells called polyribosomes show protein synthesis in action, as seen in this micrograph. Ribosomes (blue) move along a strand of messenger RNA (magenta), reading its encoded contents and assembling strands of peptides (green). Credit: Elena Kiseleva/Science Source
This article was originally published on the Quanta Abstractions blog.