As space exploration geared up in the 1960s, scientists were faced with a new dilemma. How could they recognize life on other planets, where it may have evolved very differently—and therefore have a different chemical signature—than it has on Earth? James Lovelock, father of the Gaia theory, gave this advice: Look for order. Every organism is a brief upwelling of structure from chaos, a self-assembled wonder that must jealously defend its order until the day it dies. Sophisticated information processing is necessary to preserve and pass down the rules for maintaining this order, yet life is built out of the messiest materials: tumbling chemicals, soft cells, and tangled polymers. Shouldn’t, therefore, information in biological systems be handled messily, and wasted? In fact, many biological computations are so perfect that they bump up against the mathematical limits of efficiency; genius is our inheritance.
DNA
DNA stores information at a density per unit volume exceeding any other known medium, from hard disks to quantum holography.1 It’s so dense that all the world’s digital data could be stored in a dot of DNA the weight of eight paper clips. This remarkable storage density is paired with an equally remarkable reading mechanism. A developing embryo must self-direct the rapid division, migration, and specialization of its constituent cells based on the information stored in DNA. Cells diverge from one another to grow in different ways, depending on their position in the embryo. This means that precise control of gene expression is necessary both in space and in time. Even minor errors could spell death or deformity for the organism.
The question is, how quickly and effectively can spatial information be communicated in order for development to unfold properly? Alan Turing, the father of modern computing, was fascinated by the idea that life might be reducible to mathematical laws, and tackled this question in the early 1950s. He predicted that the spatial patterning of tissues during embryonic development could be controlled through the concentration of chemical signalers, called morphogens. He derived equations showing that interactions between morphogens that activate and inhibit one another’s gene expression could set up standing waves of morphogen concentration, and control embryonic patterning. With just four variables—production and degradation rates, diffusion rate, and interaction strength—Turing could reproduce biologically plausible, self-directed pattern formation.
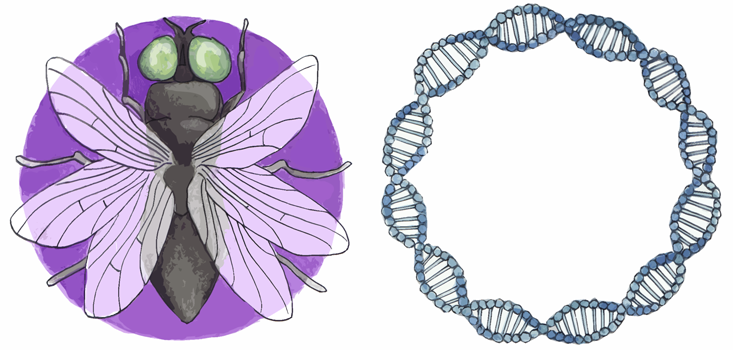
His prediction of a chemical system of information management was proven true decades later with the first discovery of a morphogen in fruit flies, called bicoid (though his equations proved to be too simple). Cells in a fruit fly embryo with high bicoid concentration become the fly’s head, and those with lower concentration become its body. What is remarkable, however, is how quickly and accurately these cells differentiate. In 2007, a team led by Princeton biophysicist Thomas Gregor measured the concentration gradient and diffusion rate of bicoid in fruit fly embryos. They estimated that it would take around two hours for embryonic cells to measure the morphogen concentration with enough precision for adjacent cells to mature differently (measurement precision increases with measurement time). But this is nearly the entirety of the fruit fly’s developmental period, from fertilization to cellularization. The embryo was developing faster and more precisely than should have been possible.
Gregor proposed that multiple cells could be sharing information with each other using a second signaling chemical: One candidate is a morphogen called hunchback. This would allow them to compute a spatial average of bicoid concentrations, rather than relying entirely on individual readings. The averaging process would be less susceptible to variation and noise, and would allow the required pattern accuracy to be achieved in about three minutes, rather than two hours.
The process is not just ingenious, but efficient. To find out how efficient, physicist William Bialek used nucleus-by-nucleus measurements of morphogens in fruit-fly embryos to study how closely hunchback concentrations followed changes in bicoid concentrations. He found that the fidelity of this information transfer was 90 percent of the theoretical maximum.
A video interview with the author, Kelly Clancy.
Fractal Networks
When an embryo develops into an adult, it confronts a new information processing challenge. Its body must learn how to sample and integrate both external and internal information. Odorant molecules signal a predator, prompting a rodent to flee. The setting sun triggers melatonin release, causing a cascade of effects throughout the body, inducing sleep. Internal chemoreceptors measure high carbon dioxide levels, causing an athlete to breathe faster. Information from microscopic measurements, sampled by disparate sensors, must be integrated and used to make macroscopic decisions that are efficiently communicated back out to relevant body parts.
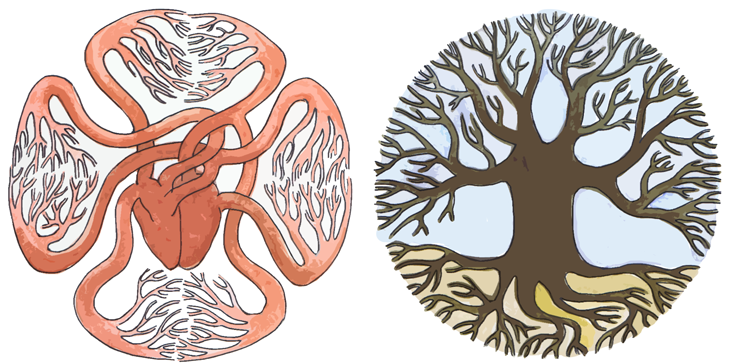
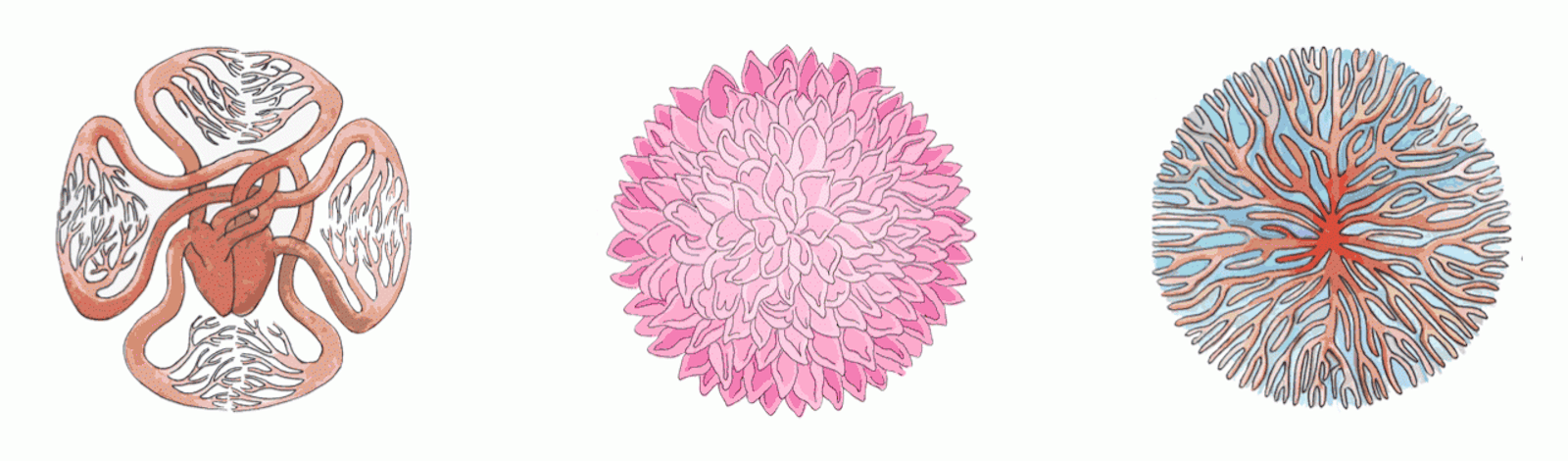
Here, too, nature has discovered an optimal solution, whose details can be read in the shapes of our anatomical structures. One of the earliest observations of a mathematical relation in biology was that almost every living creature in every ecosystem on the planet, spanning more than 20 orders of magnitude in size from uni- to multi-cellular, has features that scale with the mass of the organism, including heart rate, life span, aorta size, tree trunk size, and metabolic rate. This is called allometric scaling.
A rule of thumb common in the halls of Intel was programmed into our bodies eons ago.
Strangely, the relationship between these features and an organism’s mass usually follows a power law. That means, for example, that metabolic rate depends on the organism’s mass, raised to some constant power. Even stranger, the constant for the majority of these features is some multiple of one fourth. Physicist Geoffrey West has argued that this scaling results from the ubiquity of fractal-like networks within organisms. Fractals are structures that have similar structure at very different spatial scales—one familiar example is the vasculature of a leaf, whose branching geometry is replicated in miniature across the leaf’s surface. They can be found in a dizzying variety of systems, including our nervous system, blood vessels, capillaries in plant leaves, lung bronchioles, the calyxes of kidneys, neurons, root systems, and mitochondrial cristae.
Why would nature use fractal geometry so regularly? Mathematically, fractals are interpreted as having a fractional dimension higher than the space they reside in: A fractal drawn on a two-dimensional sheet of paper, for example, has a higher dimension—say, 2.1. This is a useful feature, allowing nature to pack some part of a fourth dimension into three-dimensional space.
To see why it might be useful, we can turn to a well-known result in the semiconductor industry, called Pollack’s Rule. It states that the speedup achievable by adding computing elements to a processor is proportional to the number of added elements, raised to the power of (1-1/D), where D is the dimension in which the processors are arranged. A three-dimensional arrangement of, say, 100 processors will give a greater boost than a two-dimensional arrangement of those same processors, due to reduced signal latency. The same kind of speedup can be achieved in our nervous and circulatory systems with a D of greater than three. A rule of thumb common in the halls of Intel was programmed into our bodies eons ago; the fractal arrangement of our biological sensors and the nerves that connect them are designed for speed.
The Brain
The brain represents the height of biological information processing and transmission. But for years, it’s been considered a noisy instrument. Neurons code information with binary electrical events called spikes, shocking one another like tiny batteries. Once a neuron receives enough input that its voltage passes a certain threshold, it will spike in turn, sending its signal to downstream neurons. Fluctuations in the summed output of the brain’s 100 billion neurons, as measured by an electroencephalogram, or EEG, are much smaller than the amplitude of individual spikes, and were long thought to be epiphenomenal: pointers to various brain states, but not an information-carrying signal in itself. In other words, noise.
Slow oscillations in the brain are coherent over large anatomical distances, in the same way you can hear the bass from your neighbor’s sound system better than the treble.
Yet, as we saw with genetic expression, interactions among many discrete elements can be incredibly useful. It’s becoming clear that the so-called noise seen in an EEG is actually an important signal, and one that is crucial to neural computation. The fluctuation of the summed output of the brain’s neurons feeds back to affect the resting potential of those neurons, bringing them closer to or farther from spike threshold. Slow oscillations (as found in drowsy or sleeping animals) are coherent over large anatomical distances, in the same way you can hear the bass from your neighbor’s sound system better than the treble. This is how the brain may be facilitating communication between disparate brain areas: If an individual spike is timed with the crest of the slow amplitude fluctuation, it will have a bigger effect downstream than if it is timed with a trough. Neurons are somehow coordinating themselves to generate self-directed, global feedback that serves to adaptively boost even sub-detectable signals: No informational signal is wasted.
In fact, it is an open question whether the idea of “waste” can even be properly defined in the context of the brain. Theoretical neuroscientist Tony Bell argues that the hallmark of biological computation is that information flows across multiple hierarchies of complexity, bubbling up from microscopic to macroscopic levels and back down again.
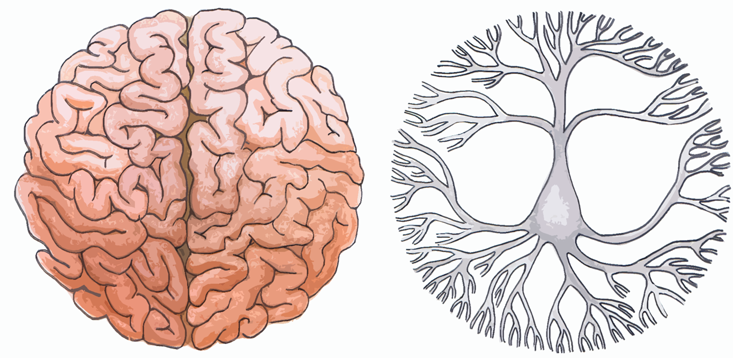
Simpler structures (like neurons) communicate with more complex structures (like areas of the brain), and vice versa. Computations are occurring on and between every level, from molecules to cells to organisms as a whole. There is a good deal of evidence that even the subcellular compartments within neurons are doing their own computations. Most recently, a study1 published in Nature in October 2013 showed that activity within dendrites (a compartment representing a neuron’s “input”) shapes the information processed by that neuron. Given this multi-level information flow, Bell argues that noise simply cannot be defined: Deviations from expectation should be interpreted as meaningful communication at another level of the complexity hierarchy.
This information flow is not restricted to the brain, and can also be observed, for example, in epigenetics. Bell says this kind of dynamic multidirectional information flow is what defines life. Artificial intelligence, when it arrives, may not be built from silicon, but repurposed biology. Before we get there, though, we will need a vastly different approach to science in order to understand the higher-order, emergent capabilities of nature’s self-organizing structures. While biomimicry holds incredible promise, perhaps in so assiduously copying the physical design of systems we are missing the point: Form is transient in Nature, cast to meet shifting needs in a given niche. Determining the rules by which biology struck upon these ingenious solutions in the first place will spark the true revolution.
Kelly Clancy studied physics at MIT, then worked as an itinerant astronomer for several years before serving with the Peace Corps in Turkmenistan. She is currently a National Science Foundation fellow studying neuroscience at UC Berkeley.
Reference
1. Smith, S., et al Dendritic spikes enhance stimulus selectivity in cortical neurons in vivo. Nature 503, 115-120 (2013).