In 1971, microbiologists examining yeast cells discovered strange, rogue fragments of RNA that turned out to be viruses. These “narnaviruses” (a portmanteau of “naked RNA viruses”) had several odd properties. They were tiny—essentially a single gene encoding an enzyme that helped the virus make copies of itself. Moreover, unlike other single-stranded RNA viruses like Ebola and influenza, they had no “capsid” shell enclosing their genetic material, leaving them exposed and restricting them to their host cells.
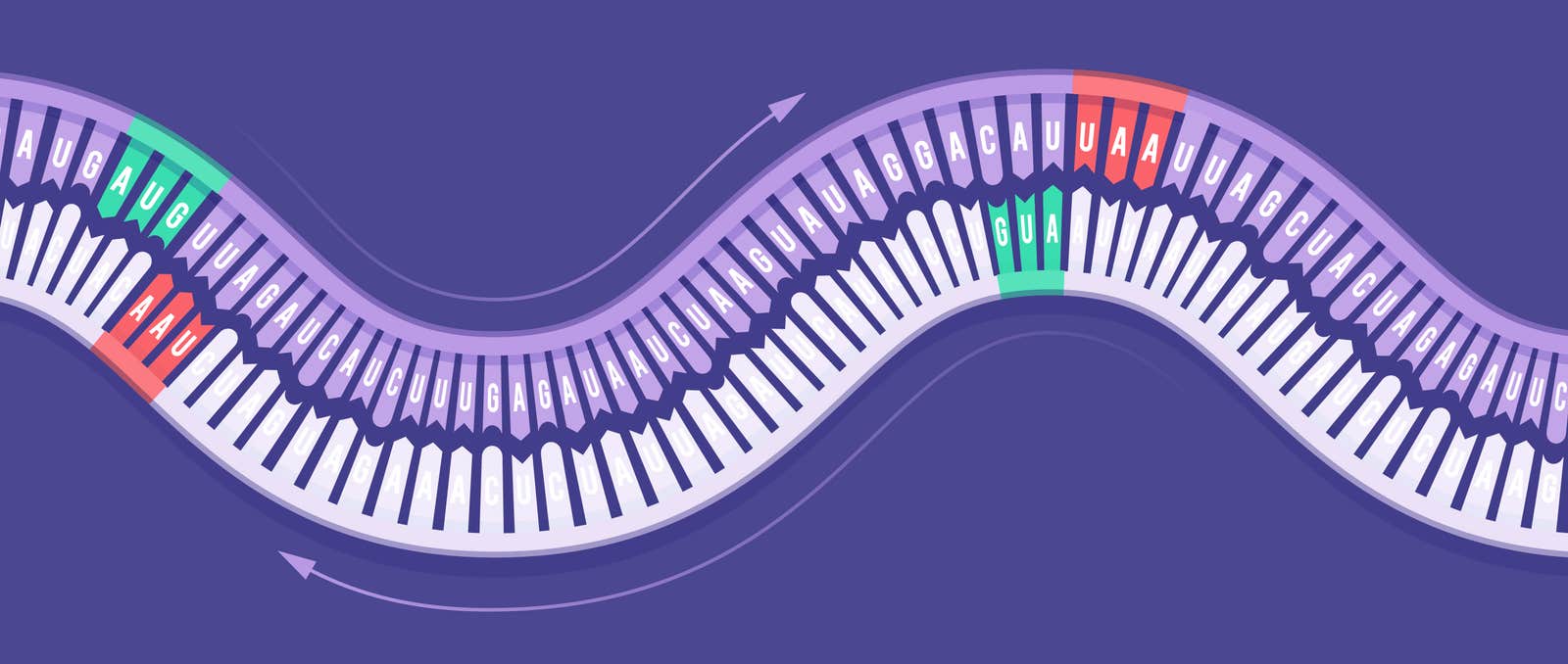
Strangest of all, narnaviruses could be read backward.
Normally, the sequence of nucleotide bases that comprise a gene—in RNA, these are adenine, cytosine, guanine and uracil, abbreviated A, C, G and U—only makes sense when protein-making organelles called ribosomes decode the message starting in one place and reading in one direction. Occasionally, sections of a genome will have overlapping sequences that code for different proteins. But in narnaviruses, the entire genome is an overlapping sequence: It can be read in its “reverse complementary” orientation. That is, the RNA is like an ambigram, a stylized script that still says something when flipped upside down.
“It’s always very surprising and interesting to find these examples that push the limits of our current conception of how information is encoded,” said Harris Wang, a systems biologist at Columbia University.
What the narnaviruses’ genome actually says in the opposite direction, and why the genome goes both ways, has been a puzzle. Now, findings by researchers in California point to an unexpected answer. The ambigrammatic property of narnaviruses may be a clever mechanism of self-preservation, one that could significantly expand the picture of viral evolution and suggest novel approaches to gene therapy.
“I think the story of these ambigrammatic viruses is going to have some legs,” said Michael Wilkinson, a physicist at the Open University in England and the Chan Zuckerberg Biohub, a life sciences research institute in the Bay Area where the research took place.
An extensive set of overlapping genes isn’t the norm because of how RNA and DNA convey information. They spell out their protein-making instructions in a sequence of “codons”—three-letter words such as CGA. Each codon tells a cell to either synthesize an amino acid (a building block of a protein) or end protein synthesis.
There are three ways to read an RNA strand, depending on which letter is interpreted as the start of a codon. But usually only one “open reading frame” makes sense. The other two have stop codons in the wrong places, rendering the interrupted genetic fragments nonsensical and incapable of forming functional proteins.
Like a record played backward, the RNA strand’s reverse complement usually doesn’t make any sense either. The reverse complement is the strand that forms when the RNA replicates—a process in which its nucleotide bases pair up with complementary nucleotides found floating in the host cell. Each A finds a U, each U finds an A, and C’s pair with G’s, so that the partners form a complementary strand, a template for making future copies of the original. The three reading frames on the template are also typically illegible, littered with accidental stop codons.
But RNA viruses are “some of the fastest evolving, most diverse replicating things in our universe,” said Joseph DeRisi, a biochemist at the University of California, San Francisco and one of the Biohub investigators. The viruses sometimes evolve overlapping nucleotide sequences that simultaneously encode multiple proteins or achieve some additional regulatory function. Although the vast majority of known overlaps appear in the same direction, as two offset open reading frames, in rare cases, most notably in HIV, the overlapping frame occurs on the RNA’s reverse complement instead.
Narnaviruses fit into that second category, but what sets them apart is how monstrously long their ambigrammatic sequence is, encompassing almost the whole genome. “It completely blows previous examples out of the water,” said the virologist David Karlin of the University of Oxford. The feature is so restrictive—handcuffing the potential evolution of the forward sequence to that of the reverse—that researchers have suspected since the 1970s that it must hold some unknown advantage.
In a preprint posted online in 2019, Andrew Firth, a virologist at the University of Cambridge, and collaborators tested various mundane explanations of the ambigrammatic property and ruled them all out. “Our conclusion is that the reverse open reading frame is functionally important,” Firth said. “We still have no idea why.”
The Biohub team reported further evidence for the feature’s significance in Scientific Reports in November 2019. First, when they investigated the genetic relationships among dozens of narnavirus species (not all of which are ambigrammatic), they found that the overlapping sequences had been gained and lost throughout their evolution. “It’s really a feature that has evolved at least twice, maybe three or more times,” said co-author Greg Huber, a biophysicist.
The researchers observed that the opposing reading frames in ambigrammatic narnaviruses were always aligned, with codon boundaries perfectly matched up. They realized that this alignment allows stop codons to disappear from the reverse sequence over the course of evolution without ruining the replication enzyme encoded by the forward strand. That is, anytime a codon in the forward sequence leaves the reverse complement with a stop, the forward codon could in theory be replaced with a “synonym” codon that translates into the same amino acid, removing the stop on the reverse complement without repercussions.
That doesn’t work when the forward and reverse reading frames are staggered. For such a long overlap, “there’s really only one way you could ever do this … and the narnaviruses use that solution,” DeRisi said. “That in turn suggests that this is not a random boo-boo on evolution’s part.”

The researchers suspected that the ambigrammatic sequence might be a means of optimizing the virus’s coding efficiency, as with other known overlapping genes. The reverse sequence doesn’t seem to encode a known protein, but it might instead regulate the gene responsible for replication or help make its protein production more efficient.
But then in late 2019, a group of Biohub researchers made a surprising discovery, one that “points to the possibility that the explanation of the ‘why’ is a little bit different,” Wilkinson said.
The researchers were analyzing the genetic material found in crushed-up mosquito samples, which have been known to contain narnavirus RNA. As expected, many ambigrammatic narnavirus genomes cropped up in the data. More surprisingly, the scientists found that cells containing the narnavirus RNA often held a second, shorter mystery RNA fragment as well. Against all odds, this shorter fragment was completely ambigrammatic too. Further work, described in a preprint, revealed that the short fragment shared an evolutionary history with the original narnavirus gene. “These results call into question what a narnavirus is in the first place,” Huber said. “It could be that a narnavirus is not simply this single stretch of RNA, but rather a somewhat more nebulous construct.”
Amy Kistler, an infectious disease researcher at the Biohub and one of the co-authors, said the multiple parts suggest that the ambigrammatic property “might reflect something about the virus—the way it replicates, the way it hijacks the cellular machinery to propagate itself in the cell.”
One leading hypothesis is that the second piece of RNA affects ribosomes, the structures responsible for translating RNA into chains of amino acids. The RNA strand might code for a protein that prevents ribosomes from detaching from both narnavirus sequences. If the ribosomes are unable to pull free of the RNA, they will accumulate until they cover the genetic material.
This would essentially camouflage the narnavirus, making it look like part of the host cell and disguising it from cellular processes that might otherwise degrade it. Ordinarily, the ribosomes would only build up along the forward strands, because the reverse complement—the viral RNA’s replication template—would have stop codons and wouldn’t look like a translatable sequence that the ribosomes should attach to. For the narnavirus’s reverse complement to also develop a ribosome coat, it would have to be ambigrammatic.
The researchers therefore posit that the ambigrammatic property is actually a protective mechanism that the capsid-less viruses have evolved to elude the host cell’s defenses.
If the virus is indeed coopting the machinery of its host cell to hide itself, “it might be something that will eventually be found to exist more generally,” Wilkinson said. “This could be a pointer to a new class of viruses.” Karlin, who is not involved in the research, agrees that the findings could be a glimpse of “a new continent of biology.”
The camouflage trick, if confirmed, could help scientists expand their gene editing toolkit. For instance, ribosome coverings might be used to artificially manipulate gene expression in novel ways. More immediately, the researchers say, this kind of ambigrammatic feature could be applied to greatly increase the payload of certain gene therapies.
The team is now performing additional experiments, spearheaded by Hanna Retallack, a graduate student in DeRisi’s lab, in hopes of fleshing out their theory and testing additional hypotheses. “I feel cautiously optimistic that there is something fundamentally quite new here,” Wilkinson said.
Update: February 13, 2020
The article was updated to add a link to the Biohub team’s new paper, an analysis of genetic (including viral) material found in mosquito samples.
Lead image: In narnaviruses, the genome’s “reverse complement” — the strand of complementary nucleotides used in replication — is legible as well. Credit: Lucy Reading-Ikkanda/Quanta Magazine