Proteins are the workhorses of all living creatures, fulfilling the instructions of DNA. They occur in a wide variety of complex structures and carry out all the important functions in our body and in all living organisms—digesting food, building tissue, transporting oxygen through the bloodstream, dividing cells, firing neurons, and powering muscles. Remarkably, this versatility comes from different combinations, or sequences, of just 20 amino acid molecules. How these linear sequences fold up into complex structures is just now beginning to be well understood.
Even more remarkably, nature seems to have made use of only a tiny fraction of the potential protein structures available—and there are many. Therein lies an amazing set of opportunities to design novel proteins with unique structures: synthetic proteins that do not occur in nature, but are made from the same set of naturally-occurring amino acids. These synthetic proteins can be “manufactured” by harnessing the genetic machinery of living things, such as in bacteria given appropriate DNA that specify the desired amino acid sequence. The ability to create and explore such synthetic proteins with atomic level accuracy—which we have demonstrated—has the potential to unlock new areas of basic research and to create practical applications in a wide range of fields.
No technology can beat the remarkable precision with which proteins carry out their unique and beautiful functions. The methods of protein design expand the reach of protein technology, because the possibilities to create new synthetic proteins are essentially unlimited.
This could not be a more exciting time for protein design.
If we were unable to predict the structure that results from a given sequence of amino acids, synthetic protein design would be an almost impossible task. There are 20 naturally-occurring amino acids, which can be linked in any order and can fold into an astronomical number of potential structures. Fortunately the structure prediction problem is now well on the way toward being solved by the Rosetta protein modeling software.
Our research team has already revealed the structures for more than a thousand protein families, and we expect to be able to predict the structure for nearly any protein within a few years. This is an important achievement with direct significance for basic biology and biomedical science, since understanding structure leads to understanding the function of the myriad proteins found in the human body and in all living things. Moreover, predicting protein structure is also the critical enabling tool for designing novel, “synthetic” proteins that do not occur in nature.
Take protein logic systems, for example. The brain is a very energy-efficient logic system based entirely on proteins. Might it be possible to build a logic system—a computer—from synthetic proteins that would self-assemble and be both cheaper and more efficient than silicon logic systems? Naturally occurring protein switches are well studied, but building synthetic switches remains an unsolved challenge. Quite apart from bio-technology applications, understanding protein logic systems may have more fundamental results, such as clarifying how our brains make decisions or initiate processes.
Proteins can also be catalysts for clean energy and medicine. Protein enzymes are the most efficient catalysts known, far more so than any synthesized by inorganic chemists. Part of that efficiency comes from their ability to accurately position key parts of the enzyme in relation to reacting molecules, providing an environment that accelerates a reaction or lowers the energy needed for it to occur. Already we have produced synthetic enzymes that catalyze potentially useful new metabolic pathways. These include: reactions that take carbon dioxide from the atmosphere and convert it into organic molecules, such as fuels, more efficiently than any inorganic catalyst, potentially enabling a carbon-neutral source of fuels; and reactions that address unsolved medical problems, including a potential oral therapeutic drug for patients with celiac disease that breaks down gluten in the stomach and other synthetic proteins to neutralize toxic amyloids found in Alzheimer’s disease.
New super-strong materials could arise from synthetic protein research. A potentially very useful new class of materials is that formed by hybrids of organic and inorganic matter. One naturally occurring example is abalone shell, which is made up of a combination of calcium carbonate bonded with proteins that results in a uniquely tough material. Apparently, other proteins involved in the process of forming the shell change the way in which the inorganic material precipitates onto the binding protein and also help organize the overall structure of the material.
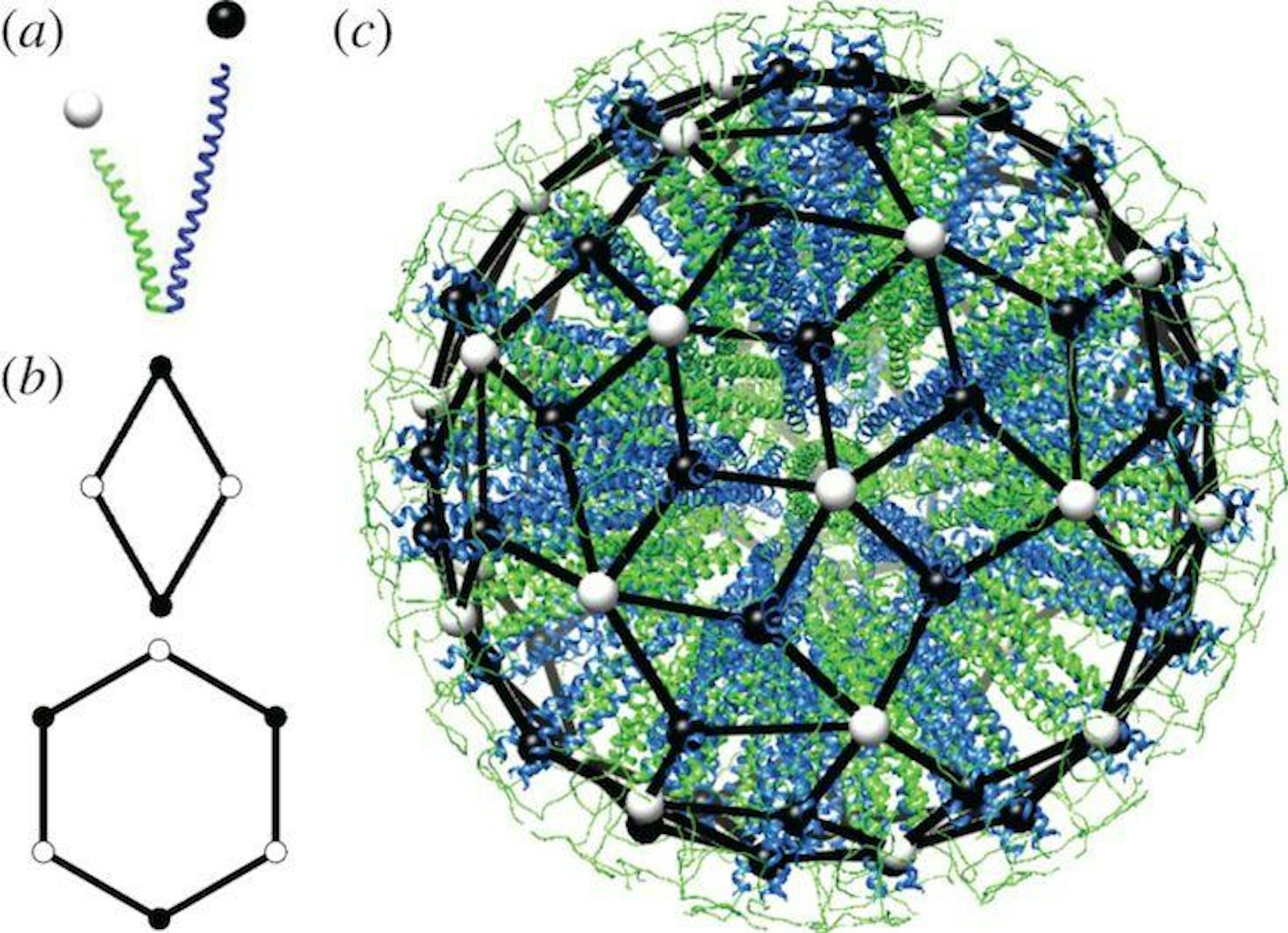
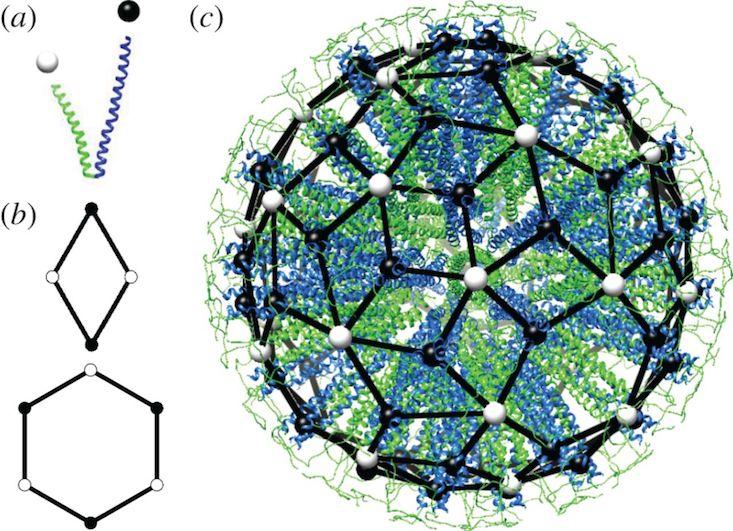
Synthetic proteins could also help advance targeted therapeutic delivery systems. Self-assembling protein materials make a wide variety of containers or external barriers for living things, from protein shells for viruses to the exterior wall of virtually all living cells. We have developed a way to design and build similar containers: very small cage-like structures—protein nanoparticles—that self-assemble from one or two synthetic protein building blocks. We do this extremely precisely, with control at the atomic level. These self-assembling particles are a completely new way of delivering drugs to cells in a targeted fashion, avoiding harmful effects elsewhere in the body. Other nanoparticles might be designed to penetrate the blood-brain barrier to treat brain diseases. More fundamentally, synthetic proteins may well provide the tools that enable improved targeting of drugs and other therapies, as well as an improved ability to bond therapeutic packages tightly to a target cell wall.
In addition to drug delivery, self-assembling protein nanoparticles are a promising foundation for the design of vaccines. By displaying stabilized versions of viral proteins on the surfaces of designed nanoparticles, we hope to elicit strong and specific immune responses in cells to neutralize viruses like HIV and influenza. We are currently investigating the potential of these nanoparticles as vaccines against a number of viruses. The thermal stability of these designer vaccines should help eliminate the need for complicated cold chain storage systems, broadening global access to life saving vaccines and supporting goals for eradication of viral diseases. The ability to shape these designed vaccines with atomic level accuracy also enables a systematic study of how immune systems recognize and defend against pathogens. In turn, the findings will support development of tolerizing vaccines, which could train the immune system to stop attacking host tissues in autoimmune disease or over-reacting to allergens in asthma.
The opportunities for the design of synthetic proteins are endless, with new research frontiers and a huge variety of practical applications to be explored. In effect, we have an emerging ability to design new molecules to solve specific problems—just as modern technology does outside the realm of biology. This could not be a more exciting time for protein design.
A funding level of $100M over five years would propel protein design to the forefront of biomedical research, supporting multiple and parallel collaborations with experts worldwide to arrive at breakthroughs in medicine, energy, and technology, while also furthering a basic understanding of biological processes. Current funding is unable to meet the demands of this rapidly growing field and does not allow for the design and production of new proteins at an appropriate scale for testing and ultimately production, distribution, and implementation. Private philanthropy could overcome this deficit and allow us to jump ahead to the next generation of proteins—and thus to use the full capacity of the amino acid legacy that evolution has provided us.
David Baker is the director of the Institute for Protein Design at the University of Washington.
WATCH: Siddhartha Mukherjee on how genetics is changing medicine.