You’re twisted. Sorry, but we all are. The molecules most central to life twist one way or the other. Your most famous molecule, DNA, a spiraling helix like the thread of a screw, is right-handed. The molecules encoded by your DNA, proteins, are left-handed. Even humble sugars like glucose have a twist to their shape.
Why does this handedness, called chirality after the Greek word for hand, feature in the molecules (clusters of atoms) from which all life on Earth is constructed? How and when was the left-or-right chiral twist of life’s components decided? No one knows—even though the chirality of life has been recognized for more than a century and a half.
But we do know that living organisms are exquisitely sensitive to this handedness. Feed bacteria with left-handed amino acids, and they’ll incorporate them into their proteins. Feed them right-handed amino acids, and they are likely to ignore them, even though these are the same molecules but inverted as though in a mirror.
There might be a dark biosphere—a whole ecosystem of mirror-image life forms.
Maybe we just need to accept this is how life is. It exists on one side of the looking glass. But some researchers think there could be equivalent lifeforms on the other side too—if only we could make them. They are already making mirror-image versions of proteins and nucleic acids like DNA. These molecules could be valuable drugs—all the more effective because their looking-glass structure should allow them to operate out of sight of the body’s usual defense mechanisms for breaking apart foreign molecules.
But biochemist Ting Zhu of Westlake University in Hangzhou, China, is determined to go much further than that. He’s made it his mission to create mirror-image versions of the key molecular ingredients of life. In principle, it might be possible to assemble those components into synthetic cell-like entities that can replicate and metabolize: a kind of primitive form of life, but inverted relative to every known organism, and therefore the first truly non-natural life form.
“I believe there are many alternative possibilities for life,” says Zhu. “But among all of these, there is one that we know for sure would work, and that’s the exact mirror-image version of ourselves.” No one knows how such mirror-image cells would get along with ordinary ones—maybe they’d ignore each other, or compete? Might our planet have already conducted a version of that experiment in the wild, billions of years ago when life began?

Over the past several years, Zhu and his colleagues have performed the monumental task of making, from their constituent molecular parts, mirror-image versions of the key biomolecules in living cells: DNA, its sibling RNA, and the enzymes for replicating them and translating their sequences into proteins. He hasn’t got the full set yet, but he is getting very close. Only a few components are still to be added to the basic biomolecular toolkit needed by stripped-down, minimal versions of mirror-image life.
“Ting’s work is really impressive,” says biochemist and Nobel laureate Jack Szostak at Harvard University, who was Zhu’s Ph.D. advisor. “Making a mirror-image cell with a complete mirror-image protein synthesis system is a hugely ambitious goal. But I think it’s worth trying, just to see what the hardest problems are.”
“This could be the beginning of a new form of life,” says Zhu. A lot of people think it can’t be done—but he is determined to try, just to see if it’s possible. “Indeed, it’s difficult and challenging, but that also makes it exciting, like climbing a high mountain.”
The Hand(s) of Life
Mirror-image life was first imagined by the man who discovered the chirality of life’s molecules: French chemist Louis Pasteur. In 1848, Pasteur deduced that a byproduct of winemaking called tartaric acid may crystallize in two mirror-image crystal shapes because their constituent molecules have opposite chirality. After carefully separating the two types of crystal by hand, Pasteur dissolved them separately in water and found that when polarized light passed through the solutions, the plane of polarization was rotated in opposite directions: to the left and to the right. Mirror- image molecular structures are called enantiomers, and are distinguished by the prefixes D and L, for the Latin words for right (dextera) and left (laeva). They are distinguished by the way their component atoms are arranged in space.
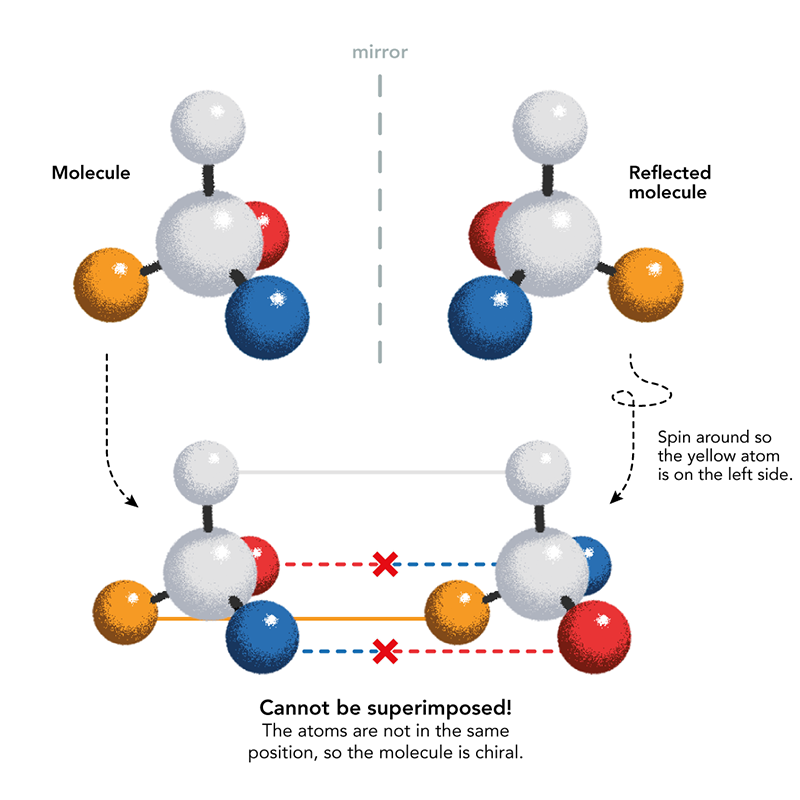
Pasteur became convinced that molecular chirality was a fundamental feature of living things: what distinguished them from the non-living world. He was wrong about that but correct that chirality is omnipresent in biomolecules. Pasteur wondered if chirality was induced by forces such as magnetism, electricity, or light, and in the 1850s he conducted a series of experiments that from today’s vantage point might look borderline cranky: crystallizing compounds in magnetic fields, growing plants in sunlight with its polarization inverted with mirrors. Those efforts never led anywhere, although recently researchers at Harvard University have proposed that magnetism in minerals might indeed have played a role in giving the earliest life its particular chirality.
But Pasteur also examined the chirality of some natural molecules and showed that the amino acid asparagine, isolated from plants (the name comes from its discovery in asparagus juice), is present in just the L-form. In 1886 the Italian chemist Arnaldo Piutti discovered the D-form of asparagine and found that, while L-asparagine is tasteless, D-asparagine is sweet. In other words, mere mirror-image inversion is enough to produce different biological effects. Such distinctions became tragically apparent when it was found in the early 1960s that the two enantiomers of the drug thalidomide, prescribed for anxiety and for morning sickness in pregnant mothers, have very different effects: One is a sedative, the other can cause serious birth defects.
Mirror Drugs
We now know that all natural amino acids in proteins are L-enantiomers. But protein-like molecules made from D-amino acids by chemical synthesis have been investigated for several decades as potential drugs.
The creation of mirror-image proteins was pioneered several decades ago by chemist Stephen Kent of the University of Chicago and his co-workers. In the early 1990s, Kent and colleagues were able
to chemically synthesize the enzyme HIV-1 protease, which the HIV virus uses to degrade other proteins. This allowed researchers to obtain the first crystal structure of the enzyme, assisting the development of drugs that combat AIDS by blocking the enzyme from working. Having acquired that facility for building proteins from scratch, in 1992 Kent’s team made a mirror-image version of HIV protease just to see what it would do. They found that the molecule would cut up only peptides also made from D-amino acids, while ignoring those made from natural L-versions.
Kent realized that such reversed proteins could be useful for drug discovery. Many drugs aim to bind to and block the action of natural proteins, and such compounds are typically found by a screening procedure: The protein target is exposed to a wide variety of compounds to see if any bind to it. Many such compounds are chiral, and a mirror-image version of the target protein could double the chances of finding a good match, because a candidate in the screening library could have the correct handedness to stick only to the mirror-image form. If so, the other enantiomer, once chemically synthesized, should stick to the natural target.
“I believe there are many alternative possibilities for life—and one will work for sure.”
D-proteins (and small fragments of them called peptides) could also be useful as drugs, for example by binding to and blocking the activity of natural proteins that are therapeutic targets. Around 80 or so normal L-peptides that do this are already on the market, but D-peptides have a distinct advantage. One of the obstacles for protein-based drugs is that natural protease enzymes quickly break them apart. But those enzymes don’t work on D-peptides, and so they “are expected to be largely invisible to metabolic degradation,” says Kent.
What’s more, D-proteins wouldn’t cause the immune system to trigger an inflammatory response, because this only happens once foreign proteins are chopped into fragments by proteases. Studies in mice have shown that D-proteins can pass harmlessly through the body before being cleared by filtration in the kidneys.
D-proteins are being explored for cancer treatment, among other things. For example, Kent’s group has explored one that can interfere with blood-vessel formation in tumors. But it’s still early days for mirror-image protein drugs. “To the best of my knowledge, none has yet been approved for use as a therapeutic,” says Kent.
Biochemist Sven Klussmann is hoping that mirror-image nucleic acids too will find uses as drugs, and he has launched two companies based in Germany to develop them. As with D-peptides, naturally occurring enzymes that degrade nucleic acids, called nucleases, cannot degrade mirror-image nucleic acids, and they are not so easily recognized by the immune response and so are less apt to trigger inflammatory reactions.
Life’s Essential Machines
While Pasteur was conducting his weird experiments on molecular chirality with magnets and light in the 1850s, he admitted, “One had to be a little mad to undertake what I am trying to do now.” That’s a sentiment Zhu might well sympathize with. His determination to build mirror-image versions of life’s key molecules from scratch, assembling their long chains piece by piece with meticulous, painstaking chemistry, goes way beyond earlier efforts to make right-handed proteins. When he began in the early 2010s, his goal must have looked crazily ambitious. But many are astonished at the progress he has made. The work “is fascinating, pushing the limits,” says Klussmann.
A bare-bones form of mirror-image life would need just a few key molecular ingredients—a little biology primer will help to explain what those are.
In cells, the enzyme DNA polymerase makes copies of DNA molecules as the cell prepares to replicate, while RNA polymerase creates so-called messenger RNA (mRNA) molecules that carry the information for making proteins. In both cases, the sequence of nucleotides made by the polymerase is determined by that in the DNA “template” strand on which the enzyme puts together the new nucleic acid. DNA nucleotides come in four varieties, distinguished by the molecules called nucleotide bases they contain: adenine (A), cytosine (C), thymine (T), and guanine (G). Each base is attached to a sugar molecule called deoxyribose and a phosphate group, which together form the backbone of the double helix on which the bases hang. Deoxyribose is a chiral molecule, and in natural DNA it is always the D-enantiomer, which is what makes the helix twist in a right-handed fashion.

The nucleotide bases can stick to one another in pairs via weak chemical bonds called hydrogen bonds. These pairs have distinct preferences because of the way the bases fit together: A sticks to T, and C to G. So the twin strands of DNA are zipped up by hydrogen bonds, and they have complementary sequences: where one has an A, the other has a T, and so on. Crudely speaking, some DNA sequences—those of the regions corresponding to genes—encode the structures of proteins, which are crumpled-up chains of amino acids. Each triplet of DNA bases, called a codon, encodes a single amino acid: This correspondence is called the genetic code.
Base pairing allows a new DNA strand to be put together on an existing one, once that part of the double helix is unzipped. The complementary pairing means that the new strand has a sequence complementary to that of the template strand: The information encoded in the sequence is preserved. All the DNA polymerase enzyme needs to do is to join together the backbone segments of new nucleotides as they dock onto the template strand.
The assembly of an RNA molecule on a DNA template strand, catalyzed by RNA polymerase, relies on the same rules of base pairing except that in RNA a base called uracil substitutes for DNA’s thymine. Once a protein-coding gene (which might typically be a few hundred bases long) has been transcribed into mRNA, the RNA molecule floats free from the DNA template strand and is translated into a protein by a large cluster of proteins and RNA molecules called the ribosome. Amino acids are brought to the ribosome attached to small RNAs called transfer RNA (tRNA), which dock onto the mRNA so that the amino acids get linked into the corresponding sequence.
The processes of information transfer involved in DNA replication, transcription, and translation comprise the so-called Central Dogma of molecular biology adduced in 1957 by British biologist Francis Crick, co-discoverer of DNA’s double-helical structure. Information goes from DNA to DNA during replication, said Crick, and from DNA to RNA to protein during transcription and translation. He also allowed for the possibility that it could pass from RNA back to DNA—a suggestion validated in the 1970s when researchers discovered that viruses that encode their genes in RNA (examples now include HIV and SARS-CoV-2) could insert them into the host DNA. This “reverse transcription” is orchestrated by a polymerase called reverse transcriptase, which itself is encoded in the viral genome.
Reversing the Parts
Zhu’s aim is to create mirror-image versions of all the components involved in enacting the Central Dogma. Then he can progress from DNA to proteins in this looking-glass world, as well as enabling DNA replication. This would give him the minimal set of biomolecules needed for putting together an inverted life form.
His first target, attained in 2016 while he was based at Tsinghua University in Beijing, was an inverted DNA polymerase. Wisely, he chose to synthesize the smallest such protein known, called African swine fever virus polymerase X. To make the inverted enzyme, Zhu’s team used now well-established chemical methods for joining amino acids one by one, although it was challenging to apply them to such a long chain. A mechanical engineer by training, Zhu learned his chemical craft under the expert guidance of Szostak, who has long pursued the challenge of making rudimentary synthetic living entities called protocells from scratch.
Once Zhu’s team had put the enzyme together, they verified that it was able to complete a 12-base strand of L-DNA on an 18-base template strand, filling in the remaining six nucleotides. The enzyme did that job rather slowly compared with natural polymerases, however, taking about four hours: Because the African swine fever virus polymerase X is so small, it isn’t terribly efficient. Given even longer—a day and a half—the enzyme was able to extend such a 12-base “primer” strand all the way along a 56-nucleotide template. Better still, the mirror-image polymerase worked for making RNA too, transcribing L-RNA on an L-DNA template.
“The devil is in the details,” he says cautiously, “and there are a lot of devils.”
It would be useful to use such a mirror-image DNA polymerase to amplify strands of L-DNA—that is, to make copies from which it can make further copies, boosting the number of strands exponentially. That’s what happens in the process called the polymerase chain reaction (PCR), ubiquitous in genetic biotechnology for making lots of DNA from tiny samples. (It’s the process used in the PCR test for COVID-19, where tiny amounts of viral genome in swab samples are amplified so that they can be identified.) To conduct PCR, the double-strands made by a DNA polymerase are separated so that each strand can act as a template for further replication, and the cycle is repeated many times. The strands are separated by warming them up, which unzips the hydrogen bonds holding them together. This heating is apt to destroy most polymerase enzymes, but standard PCR uses one taken from a thermophilic (“heat-loving”) bacterium that lives in hot thermal springs, which is more resistant to heat.
As his next target for mirror-image biochemistry, Zhu chose a DNA polymerase called Dpo-4 from a heat-tolerant microbe, Sulfolobus solfataricus. The researchers made it in 2017 and showed that indeed it could be used for looking-glass PCR. In 2019, Zhu’s team made a mutant form of the polymerase that could not only transcribe L-RNA from L-DNA but also reverse-transcribe it, writing an RNA sequence back into DNA: another link of the information-transfer network in the Central Dogma.
In 2022, Zhu and colleagues synthesized the mirror-image form of the massive 883-amino-acid protein RNA polymerase used by the T7 bacterial virus, a widely used enzyme for transcription because of its efficiency and accuracy. This achievement, says chemical biologist Richard Payne of the University of Sydney, “represents a monumental feat of total chemical synthesis and a pivotal breakthrough on the journey toward mirror-image life.”
With nucleic-acid polymerization under his belt, Zhu moved on to the biggest challenge for a mirror-image Central Dogma: a reversed ribosome capable of translating L-RNA into D-proteins. The ribosome is not a single enzyme but a huge piece of biochemical machinery, with many component parts. In the ribosomes used by the common gut bacterium Escherichia coli, for example, there are around 55 proteins and three ribosomal RNAs.
Zhu began working toward this goal in 2019, and the following year his group at Tsinghua reported that they had synthesized mirror-image versions of three key ribosomal proteins. They used their polymerases also to make a mirror-image ribosomal RNA molecule and showed that the three proteins would assemble spontaneously with this RNA into the complex found in the ribosome.
Now Zhu says that he and his team at Hangzhou have made nearly all the molecular components of the ribosome, although they have not yet published this work. How close is he? “The devil is in the details,” he says cautiously, “and there are a lot of devils.”
Life, Inverted?
If Zhu succeeds in making a mirror-image ribosome, it could be a huge boost for D-protein therapeutics, because then such proteins could be made simply by producing the corresponding mirror-image DNA—a simpler task—and transcribing and translating it.
“A working synthetic ribosomal system for making mirror-image proteins would be a practical demonstration of the power of chemical synthesis applied to enzyme protein molecules,” Kent says. “It would show that we do in fact understand the key aspects of how proteins are biosynthesized.”
Yet Zhu’s ultimate goal is not drugs but life itself: making a mirror-image protocell with all that it needs to replicate and to use its genome to generate the enzymes it needs to function and metabolize. That would arguably constitute the first genuinely artificial life form. Goodness knows what such a thing might be used for, but Kent attests this curiosity-driven work—what he calls “ambitious and excellent science”—is its own justification. “Good research does not need to be justified by practical applications,” he says.

MOUNTAIN CLIMBER: Chinese biochemist Ting Zhu is determined to create a new form of life by reversing the shape of key molecules. He agrees it’s incredibly challenging, “but that also makes it exciting, like climbing a high mountain,” he says. Photo courtesy of Ting Zhu.
Can such mirror-image life ever be made in the lab? A minimal protocell that can transcribe and translate its genome into functional proteins need only have the basic molecular ingredients encapsulated within a lipid membrane. A more ambitious goal would be an entire mirror-image bacterium, with all its nucleic acids, proteins, and sugars inverted. If such an organism is ever made, it should look and act no different from a normal bacterium. Only by zooming in on the individual molecules would the looking-glass difference become apparent.
Right now, though, all this is still fantasy. Zhu points out that no one has yet made a replicating synthetic protocell even using the normal biomolecular machinery. He thinks it makes sense to wait until that is done before trying to do it with mirror-image molecules.
Mirror-image life might also help to explore “one of the biggest mysteries surrounding the origin of life on Earth,” says Zhu—its “homochirality,” or why it uses only D-nucleic acids and L-proteins. Some think the choice was entirely random: Perhaps both enantiomers existed among the prebiotic building blocks, and some random fluctuation in the concentrations of one type got amplified by feedback processes and gained precedence. Others wonder if some tiny chiral bias might have been introduced by more fundamental factors, such as the left-right symmetry-breaking called parity violation, known to occur in nuclear processes involved in radioactive beta decay.
Having a mirror-image biochemistry might not answer the question of homochirality, but it could supply a new direction to probe: by putting both enantiomers of the biomolecules together in the same test tube and seeing what happens, especially if the molecules can mutate and evolve. Zhu says that in dilute solutions, mirror-image proteins and nucleic acids seem to ignore one another. But if they interact in more concentrated conditions, might just a single chirality emerge from the melee?
There is another possibility, too. “Is life on Earth really homochiral, or do we just not have the right tools to look for the other chiral version?,” Zhu asks. What if mirror-image life exists in some corners of the natural environment that we’ve overlooked because we don’t have the means to detect it? There might be a dark biosphere—a whole ecosystem of undetected mirror-image life forms. If so, a PCR system that can amplify the inverted DNA from such organisms could help to bring it to light.
Once he has made a working mirror-image ribosome, however, Zhu says he’ll probably leave the challenge of complete mirror-image protolife to others. At that point, he’ll be ready to move on to other projects. Besides, he has another way to explore the dreams of alternative life. “Things I can do, I’ll do in the lab,” he says. “Things I cannot do yet, I’ll write them as science fiction.” ![]()