A few years ago, James Watson, one of the co-discoverers of the double helix structure of DNA, wrote a manifesto on cancer. He attacked the pursuit of cancer drugs based on next-generation personal genome sequencing, which seeks to unravel the genetic mutations that enable cancer. “The now much-touted genome-based personal cancer therapies,” he wrote, “may turn out to be much less important tools for future medicine than the newspapers of today lead us to hope.”
Although next-generation sequencing is now affordable, and even allows us to read the genetic code of a single cancer cell, incident rates of new cases of cancer are still on the rise; and while the cancer death-rate is slightly down, most of the gains have been made in blood and immune cell cancers, like leukemia and lymphoma, not the solid-state tumors deep in our organs, which are harder to detect and treat. The use of personal genetic profiles to rethink cancer was no giant leap for mankind, and the press, Watson pointed out, began to report the hyperbole. We’re gaining “comprehensive views of how most cancers arise and function at the genetic and biochemical level,” he noted. But the vast set of tricks that cancer cells can use to escape cellular controls on their growth and migration means that the “curing” of cancers now unfortunately seems “an even more daunting objective” than it was in 1971, when Richard Nixon initiated the “war on cancer.”
Simply learning how to stop cells from becoming cancerous doesn’t seem to be a promising way to fight cancer, since hundreds of human genes are serious “drivers” that may alter multiple pathways—any chain of events that cells can readjust to alter energy use or production, or to grow and divide abnormally. Other molecules bind to DNA or to the proteins that holster it, resulting in cell types as diverse as bone, liver, or neuron—miraculous changes that can happen without ever altering the permanent code of a cell. (Each tissue or cell—no matter the type—has a unique epigenetic code that determines what genes are expressed and what type of cell it becomes.) When cells become cancerous, they may reverse these shapeshifting tricks, undergoing so-called “epithelial-to-mesenchymal cell transitions.” This turns dedicated cells—that is, cells working as a specific type—into free-floating cells. Their flexible shapes and ability to generate high amounts of the molecular fuel, ATP, allow them to achieve “anchorage independence.” They break from neighboring cells, becoming “motile,” or free to move elsewhere—to the brain, liver, or lungs, perhaps. Mutations to genes in some key pathways related to energy use or cell division can also enable cells to undergo unchecked growth or proliferation.
Mutations that cause cancer in an adult may even occur while that person was an embryo.
Most cancer treatments therefore focus on blocking or targeting the known genetic variants that enable a cell to become cancerous, grow, or proliferate. But it’s also possible to experimentally “knockdown” every gene to find which genes can be deactivated to exploit cancer cells’ vulnerabilities, and hence kill them. So, rather than target genes that enable cancer cells to grow, many cancer researchers, as Watson heralded, began to shift strategy to focus on identifying a set of genes—called “synthetic lethals”—that could be deactivated to kill a cancer cell. Synthetic lethals consist of two or more genes, of which a cell requires at least one functioning to stay alive; deactivating both critical genes leads to a cancer cell’s death.
Watson liked this: Better to fund research into identifying synthetic lethals, he argued, and even better to focus on lethals that can apply to anti-metastatic drug development, since metastasis—cancer’s spread into other parts of the body—leads to 90 percent of cancer deaths.
Identifying the genes responsible for metastasis has been confounding. There doesn’t seem to be a discernable pattern. Recent work by Bill Hahn, an oncologist at Dana-Farber Cancer Institute, in Boston, and Charles Swanton, of the Francis Crick Institute, in London, helps explain why: Metastasis-promoting mutations don’t seem to belong to any specific gene or gene family, or associate with any specific pathway, but appear to be fairly randomly distributed among both genes and pathways—and actually may occur early in cancer’s onset. For example, mutations that cause cancer in an adult may even occur while that person was an embryo.
Researchers were holding out hope, though, that they could make progress on the problem by focusing on not how to stop cancer from forming, but on how to detect cancer cells’ fatal weaknesses once they arise.
One of these strategies depends on using a “screen.” RNAi screens, in particular, were developed to introduce so-called short hairpin RNA molecules. These can latch onto messenger products of genes, called messenger RNA, and initiate natural cellular processes to deactivate, or “knockdown,” its expression, stopping it from being translated into a functional protein. “CRISPR screens” were later developed to disrupt the gene itself, to stop the gene from producing messenger RNA.
In 2011, a team of researchers discovered a knockdown of the protein Brd4. This is encoded by the gene BRD4, and is critical to the maintenance of acute myeloid leukemia, a cancer that begins in the bone marrow by generating abnormal blood cells. Deactivating Brd4, along with three other genes that work in coordination, resulted in a decrease in the expression of MYC, a gene that plays a role in the cell cycle and cell growth. Although MYC is technically “undruggable”—its role as a transcription factor means it resides inside the cell’s nucleus and is tough to reach—by deactivating these other four genes “upstream” of MYC, the team can actually control and reduce the expression of the protein Myc (encoded by MYC) which results in programmed cell death, or apoptosis. In other words, deactivating a few genes that can subsequently reduce expression of MYC could be used to kill the cancer cells of an incurable form of acute myeloid leukemia. The finding was especially promising, since deactivating MYC may be a key to stopping many cancers, including metastatic forms.
Myc’s expression is activated through two major pathways. One is the JAK-STAT pathway, and the other is the EGFR-Ras-Raf-MEK-ERK pathway. Along with being a mouthful, it’s also a cascade of proteins, including growth signal receptors and special enzymes, called kinases, which have a role in cell division or differentiation. These can be altered through mutations to any one of those genes to initiate cancer, and turn out to be highly druggable. For instance, an inhibitor to the kinase ALK, which impedes the JAK-STAT pathway, is a main drug target of some lymphomas and non-small cell lung cancers. Indeed, small molecule drugs exist for EGFR-driven lung cancers, KRAS-transformed cells, BRAF-induced melanoma, but variations of these cancers are also drug resistant.
“When does ‘treatable’ mean ‘curable’? I’m not sure, but living five years with pancreatic cancer would be quite something.”
While a single move or target might not be sufficient to kill a cancer cell, researchers have used screens to learn the right combination of genes to achieve synthetic lethality—in effect, deploying a kind of pincer move on cancer. For example, a 2011 report published in the Journal of Clinical Oncology concluded that KRAS-mutated cells in colorectal cancer could be effectively killed when a series of products of genes in the Ephrin family, receptors that turn on kinase signaling, were deactivated. The advancement of these screens can be seen as a small vindication of Watson’s call for a shift in strategy. It offers hope that we will be able to deduce a multiple gene-target formula that can be drugged to stop a cancer rather than having to carry on what might be endless research into how a cell can become a cancer.
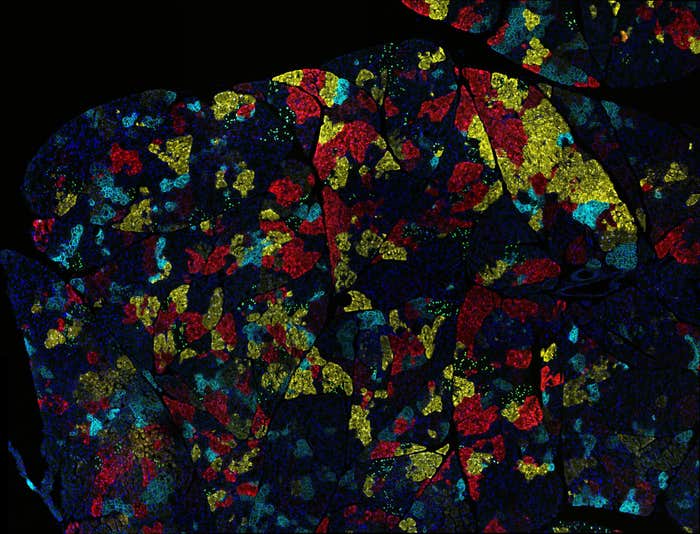
RNAi and CRISPR screens are now attracting huge interest not just at Watson’s home base, Cold Spring Harbor Laboratory, in New York, but also at the Broad Institute in Cambridge, which is conducting screens on hundreds of cell lines. The $10 million Achilles Project was set up, a decade ago, as a consortium between the Broad Institute 501c3, and now, a half-dozen industrial partners, who chipped in 20 percent of the cost in return for analysis of data on cancer cell lines. The first phase of the project included an RNAi screen on 500 cell lines, whereby researchers knocked down each gene with at least five hairpins (so approximately 90,000 hairpins in total). After the cells grew, the scientists sequenced them to discover which hairpins were depleted, concluding that those interrupted cell growth.
This past winter, Achilles project scientists completed work on 300 cell lines, using CRISPR-Cas9 to systematically knock down every gene. They’re realizing that many of the genes they knockout, resulting in killed cancer cells, are already known drug targets, so they’re doing something right. It means that they’re on stable technical ground to potentially find more targets that haven’t been described before. Other CRISPR knockdown experiments, by contrast, are challenging some established facts about cancer. This week a report on a CRISPR knockdown of the gene MELK, for example, found that it failed to stop cancer cell proliferation, even as clinical trials are underway to test inhibitors of the MELK protein.
The Broad Institute also began work with I.B.M. Watson Health, a question-answering natural-language computer system, around the same time to take the project a step further. It’s applying small molecule drugs to cancer cell lines, in combination with RNAi and CRISPR-Cas9 screens, in hopes of identifying extra genes that can be knocked out for a one-two punch. “We have a parts-list of cancer, and what we are now doing is reverse engineering the system by disrupting the function of genes,” said Hahn, who leads the project. “Anything discovered in the past 40 years in cancer research”—any gene that can be targeted to stop a cancer cell, basically—“we’ve found it. We’ve also found several hundred new things.” Hahn said those genes can’t all be targeted with a small molecule drug or antibody, but others can be: We “certainly have more leads than we can pursue.”
For example, it’s also long been known that breast and ovarian cancer cells with a hallmark mutation to a gene called BRCA—which works as transcription factor and repair enzyme—respond better to treatments that inhibit the product of a gene called PARP1, which plays a role in helping cells repair breaks in their DNA. These genes can be lethally deactivated.
Findings from the screens are also providing new leads. For example, David Barbie, an oncologist at Dana Farber, with Gilead Pharmaceuticals, is aiming to enroll patients in a study on the effects of a small molecule drug known to block the function of a kinase built by a gene called TBK1. It’s a promising new target since the knockdown of the product of TBK1 affects cancer cells harboring a mutated KRAS gene—yet another example of lethal deactivation. In effect, Barbie’s experimentally deducing the logic of the key genes a cancer cell needs to survive. David Root, head of the platform at the Broad Institute, which works on technologies such as RNAi or CRISPR-Cas9 for Achilles, puts it like this: “We are creating a hypothesis of tumor vulnerabilities so that we can get smarter about which targets are worth trying to develop.”
Watson himself, now 88, used to be pessimistic about curing cancer when gene-targeted drugs, he said, “began to fail.” But now he’s optimistic. “When does ‘treatable’ mean ‘curable’? I’m not sure, but living five years with pancreatic cancer would be quite something. I don’t want to die until I see that most cancers have become curable.”