Imagine you had a way to cure cancer that involved taking a molecule from a tumor and engineering the body’s immune cells to recognize and kill any cell with that molecule. But before you could apply this approach, you would need to be sure that no healthy cell also expressed that molecule. Given the 20 trillion cells in a human body, how would you do that? This is not a hypothetical example, but was one recently posed to our laboratory. More fundamentally, without a map of different cell types and where they are found within the body, how could you systematically study changes in the map associated with different diseases, or understand where genes associated with disease are active in our body, or analyze the regulatory mechanisms that govern the production of different cell types, or sort out how different cell types combine to form specific tissues?
We suggest the answer to these questions is to create a Human Cell Atlas that organizes cells by type and location in specific tissues. What makes this now possible is the recent development of three new tools:
- The ability to rapidly determine cell types by rapid capture, processing, and RNA sequencing of single cells. The RNA sequence reveals the genetic profile of an individual cell—identifying which genes are turned on, actively making proteins. It is in effect the zip code for cell types. Newly-invented automated tools can now process thousands of cells for sequencing per second for a low cost.
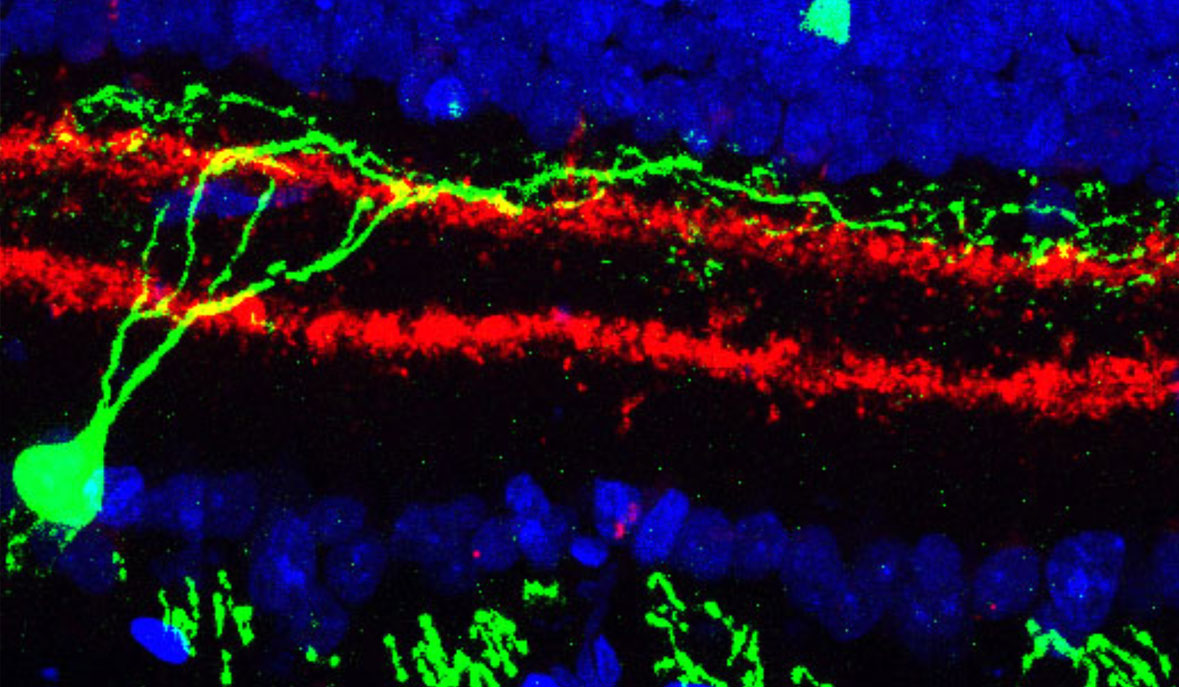
- The ability to map the location of specific cell types within living tissue at high resolution. One technique uses ion beams that scatter cell particles from specific locations in a tissue; the cell types are identified by the proteins they use and that information synthesized with the location into an image that locates the cells of interest.
- Sampling algorithms and Big Data computational techniques that enable creation of an overall cell census. These approaches are new to biology on the scale proposed here. With appropriate sampling, analyzing just 50 million cells—one for every 400,000 in the body—can give a detailed draft picture of human cell types. Big Data techniques can then be used to combine zip codes and physical locations into a unique and invaluable reference database.
Cells are the basic unit of life, yet they vary enormously. Huge quantities of new red blood cells are made every day, whereas nerve cells—especially the neurons that are the processors of the brain—are made early in life and new ones are rarely born thereafter. The types of cells also vary widely from one tissue to another. The lining of the gut contains cells that absorb nutrients, immune cells to fend off harmful microbes, and neurons—as well as cells of the beneficial bacteria that colonize us. The retina at the back of the eye functions as a kind of digital camera, capturing an image and shipping it off to the brain for analysis—and it contains more than 100 different types of neurons; one kind of neuron can be important to identify when the light is turned on, another for when the light is turned off, and so on. The T-cells of our immune systems come in different forms, depending on whether they are found in the blood, in the gut, in the mouth, or in nasal passages. Moreover, variations in specific genes that can lead to disease typically manifest themselves in specific cells, those cells where the genes would normally be active—muscular dystrophy in skeletal muscle cells, for example. Both this enormous variety from one type of cell to another, and the mix of cells from tissue to tissue are critical to the functioning of our body, but have not been fully studied or characterized.
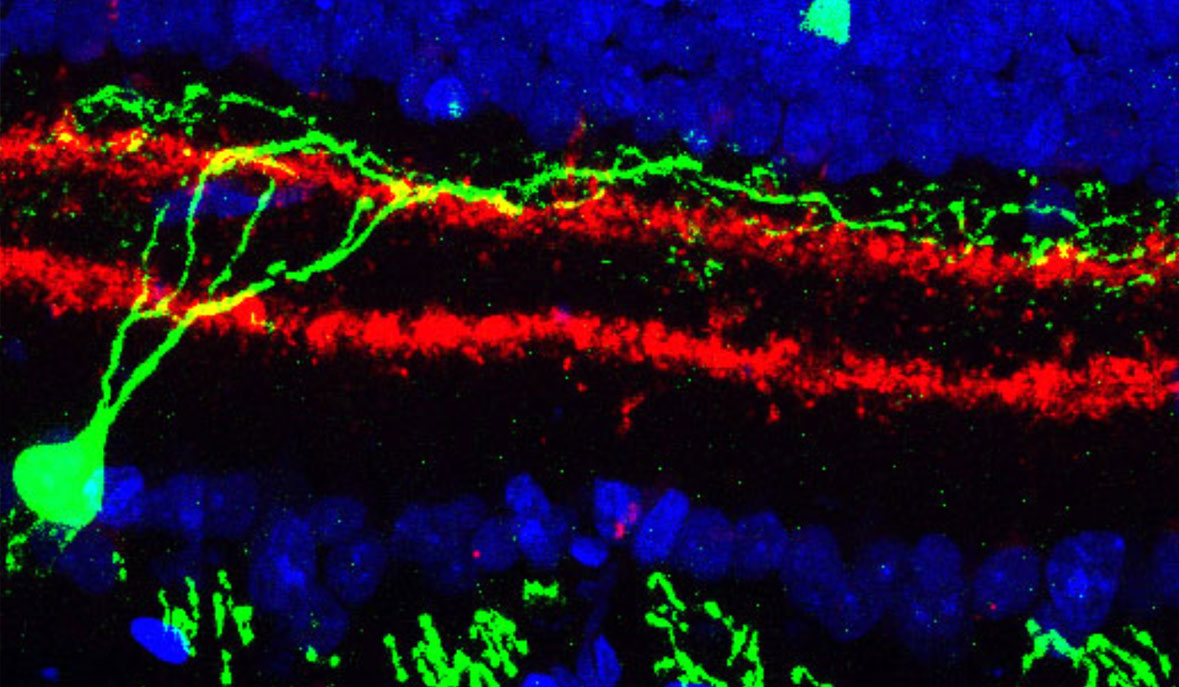
Already, in preliminary studies of the type proposed here, our lab and collaborators have discovered a completely unknown type of dendritic cells—immune cells that constitute our first line of defense against pathogens—that make up only 4 of every 10,000 cells in the blood. Another study of a particular class of T-cells associated with autoimmune diseases found subtle differences in cells taken from the gut and from the brain, changes that appear to stem from fats in the diet and that may suggest new drug targets for treating these autoimmune diseases. Analyzing tens of thousands of retina cells led to the discovery of two new cell types that have eluded decades of meticulous research.
Some 25 years ago, scientists first proposed the Human Genome Project to systematically discover all of the cellular components encoded by our genes. At the time it seemed an audacious goal, but one that proved achievable. We now propose a similar systematic effort to define the cells that underlie human health and disease.
Specifically, within five years we propose to generate a detailed first draft of a molecular atlas of cells in the human body. This Human Cell Atlas will:
- Catalog all cell types and sub-types.
- Distinguish cell states (e.g. a naive immune cell that has not yet encountered a pathogen compared to the same immune cell type after it is activated by encountering a bacterium).
- Map cell types to their location within tissues and within the body.
- Capture the key characteristics of cells during transitions, such as differentiation (from a stem cell) or activation.
- Trace the history of cells through a lineage—such as from a predecessor stem cell in bone marrow to a functioning red blood cell.
Just as with the Human Genome Project, the task is large but finite and can only be done successfully within the context of a unified project that engages a broad community of biologists, technologists, physicists, computational scientists and mathematicians.
Some factors that point toward success of the project include:
Manageable Scale:
The number of human cell types depends on the level of resolution at which they are defined. A few hundred types are often quoted, but just the blood and immune system alone may have over 300 molecularly and functionally distinct sub-types. While the number appears daunting at first, there are multiple cell “copies” of the same type, and thus this is a sampling problem. Statistical considerations and mathematical theory suggest that we can sample a manageable number of cells and still recover fine distinctions with confidence.
Sample Collection:
Experience has shown how to acquire excellent collections of human tissue samples with a well-concerted effort, even by individual labs. And, unlike genetic studies, a large number of individuals is not required. We propose to complement human sampling with limited similar studies of model organisms—primates, mice, and others—to obtain otherwise inaccessible samples and to relate knowledge from human cells to that obtained from lab experiments, for which there is extensive legacy knowledge from decades of scientific research.
Inclusive Organization:
We envision a community-wide effort that balances the need for domain-expertise in a biological system with opportunities for new technologies (more so than in past genomics projects), and yet also enables data collection that is comparable across systems. Within such a consortium, to be defined through a community process, there will be working groups for human samples, model organisms, and technology development, in addition to centralized data acquisition and management. We would expect multiple analytics efforts.
Appropriate Staging:
A Human Cell Atlas is an endeavor of new scale and type. A pilot phase that can be established quickly and serve to test alternative strategies and to evaluate the basic premises of the work would likely be particularly effective. We propose a pilot phase with a relatively sparse survey of 100,000 cells from each of 50 carefully chosen tissues from human and mouse, complemented by a much deeper survey in a few well-chosen complementary systems, such as peripheral blood and bone marrow, gut, and liver. A full-scale project, building on the pilot, would analyze more cells per tissue, additional tissues, expand work in model organisms, and deploy more measurement techniques; it could also extend analysis to disease tissues.
Having a complete Human Cell Atlas would be like having a unique zip code of each cell type combined with a three-dimensional map of how cell types weave together to form tissues, the knowledge of how the map connects all body systems, and insights as to how changes in the map underlie health and disease. This resource would not only facilitate existing biological research but also open new landscapes for investigation. A Human Cell Atlas will both provide foundational biological knowledge on the composition of multicellular organisms as well as enable the development of effective medical diagnostics and therapies.
How to Create a Human Cell Atlas
The field of genomics has substantial experience in large-scale projects such as that proposed here, but there are important differences. Genetic studies often focus on differences in the DNA between individuals, but cannot tell the critical differences between individual cells, including where the genetic differences manifest themselves. Indeed, within an individual, nearly every cell has the same DNA, but it uses (or “expresses”) only a portion of it. In contrast, a Human Cell Atlas focuses directly on the differences among cell types and is thus more diverse and complex—tracking several very different types of data—and requires more technological and computational innovation.
What makes such a project possible is very recent advances in the ability to analyze the genomic profile of a single cell. That means determining which RNA molecules it expresses from its DNA, which proteins are expressed from the RNA, and related information such as how the cell’s DNA is decorated with additional molecules that control it. With recent breakthrough technologies this can be done for large numbers of cells very quickly and inexpensively. This data characterizes which genes are active in a given cell—in effect, which proteins it produces, and what the cell does.
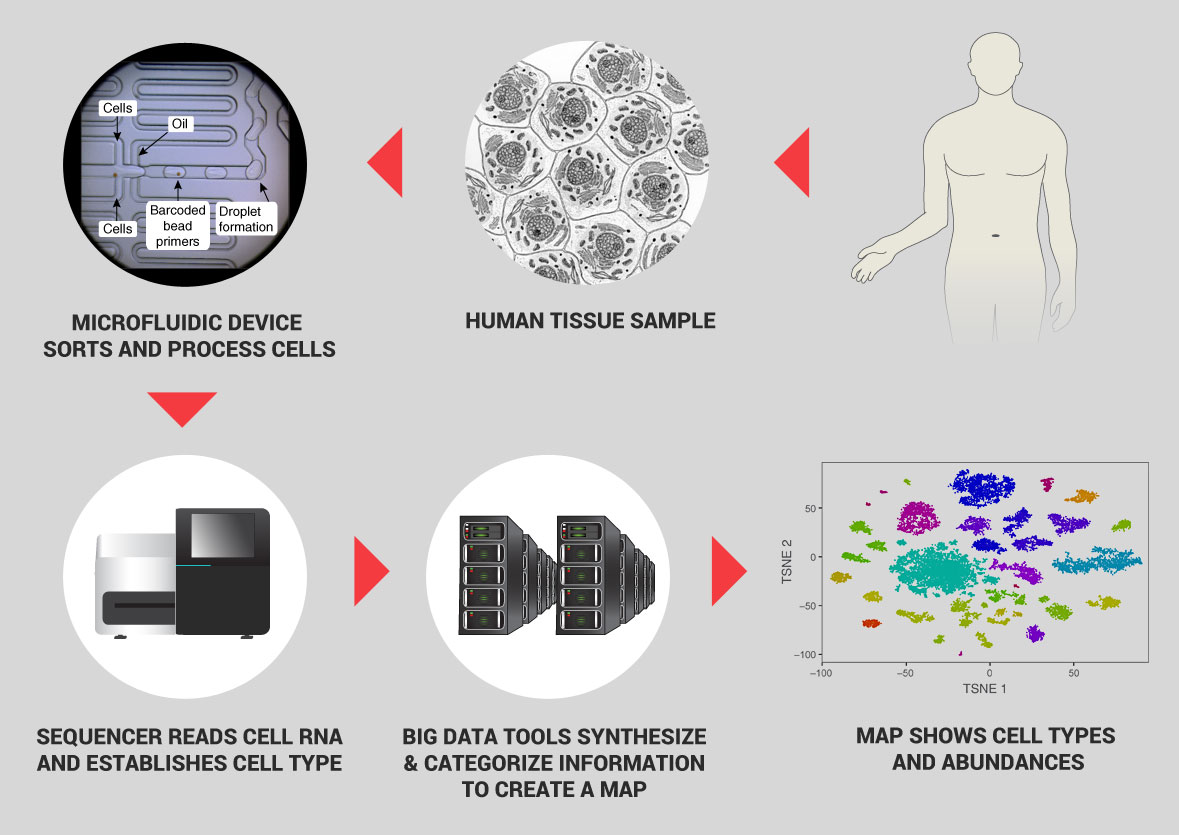
These innovations—based on advances in molecular biology, microfluidics, droplet technology, and computation—now enable massively-parallel assays that can process hundreds of thousands of cells at very low cost; we estimate a cost of about $0.17 per cell. A second emerging method of characterization involves imaging cells inside tissues at high resolution. Finally, new experimental and computational techniques couple molecular profiling (of RNA or proteins) with ion beams to high resolution spatial information about their location within a tissue or even within a cell, providing a unique characterization of the structure of tissues. We estimate that overall cost for characterizing cells by these combined methods will be $1 per cell (an estimate that assumes continuing reduction in sequencing and storage costs, and a focus on RNA measurements as the first line of characterization). We propose to analyze 50 million cells in a five year initial effort.
This initial phase of the Human Cell Atlas will also define markers for different cell types, for which antibodies and other probes can be developed to find specific cell types within a tissue. It will provide a direct view of living human tissue—removing distorting effects of cell culture on which much current knowledge is based. It will provide a way to integrate a large body of legacy data. Moreover, the Human Cell Atlas will help uncover the regulatory processes that control cell differentiation and cell interactions. Finally, and non-trivially, the project will generate standardized, tested, and broadly applicable experimental and computational methods that will be useful in many other contexts.
A level of support of $100 million over five years would support the initial organization and execution of this ambitious effort. Federal funding on this scale is unlikely, and multiple grants would not allow the integration of expertise across many groups of investigators and the complex coordination needed to ensure comparable and reproducible results. The Human Cell Atlas is thus an extraordinary opportunity for private philanthropy.