Our genetic blueprint charts the course for our life, yet we rarely achieve our full genetic potential, because of external forces that continually steer us off course. Many environmental influences are beneficial—good nutrition, education, and socialization—while others, such as malnutrition, pollution, and poverty, contribute to ill-health and the woes of humankind. We can now measure and utilize over a billion features of the genome—the sequence of DNA, epigenetic changes that turn genes on and off, and how genes interact with the biochemical machinery of cells—and that knowledge is enabling powerful new insights and therapeutic approaches. But genetics can explain less than 25 percent of most major disorders. And at present our ability to measure the complex environment in which we live and its impact on our bodies is very limited.
One measure of the potential environmental impact on health is the registry of nearly 80,000 industrial chemicals that is maintained by the Environmental Protection Agency. The interaction of each of these chemicals with living organisms can generate multiple chemical markers, which could be used to assess exposure and potential harm, if we knew what they were. One example that has generated widespread concern is the class of phthalate chemicals used as plasticizers in a wide range of products, from infant lotions and powders to credit card purchase receipts, markers from which are found in virtually every United States resident, and which have been implicated as hormone-like endocrine disrupters that can affect sexuality. Those aren’t the only powerful chemicals in consumer products—think suntan lotions and beauty creams, preservative additives in food products to extend shelf life, pesticide residues on fresh produce. Food itself generates many different metabolic chemicals found in our bodies, both directly and as a result of processing of food by our microbiome—the colony of bacteria that inhabit our gut and our skin and are very much part of who we are. Add in air pollution, workplace hazards, the markers left by allergens and disease agents and immune system reactions to them, the medicines we use—they all leave a biochemical marker. The number of such markers in our bodies is estimated to be as many as 1 million, but what they are and which ones indicate conditions or exposures harmful to health is still for the most part unknown.
As it turns out, there is no reason for knowledge of the environmental mediators of disease and health to lag so far behind that of the genome. When the concept of the exposome—the totality of our exposures from conception onward—was first put forward in 2005, it seemed an impossible challenge: How could we detect and measure a million different chemicals? But just as a single sample of blood contains the core of our genetic data (in the DNA within white blood cells), so that same blood sample contains hundreds of thousands of biochemical markers. And because of advances in high resolution mass spectrometry and high throughput screening, it is now possible to identify, catalog, and understand each marker, each constituent of the exposome, and link it to the process or environmental factor that produced it. Mapping and annotating these biochemical markers, creating reference databases that define the human exposome and to which individual profiles can be compared, using big data techniques to correlate genetic and environmental factors—all of this portends a revolution in how we understand the complex chemistry of life. That knowledge in turn is likely to lead to more specific insights into how environmental factors affect human health and how we might intervene to protect and enhance it. Within a decade, potentially, a truly precision approach to personalized medicine could be possible.
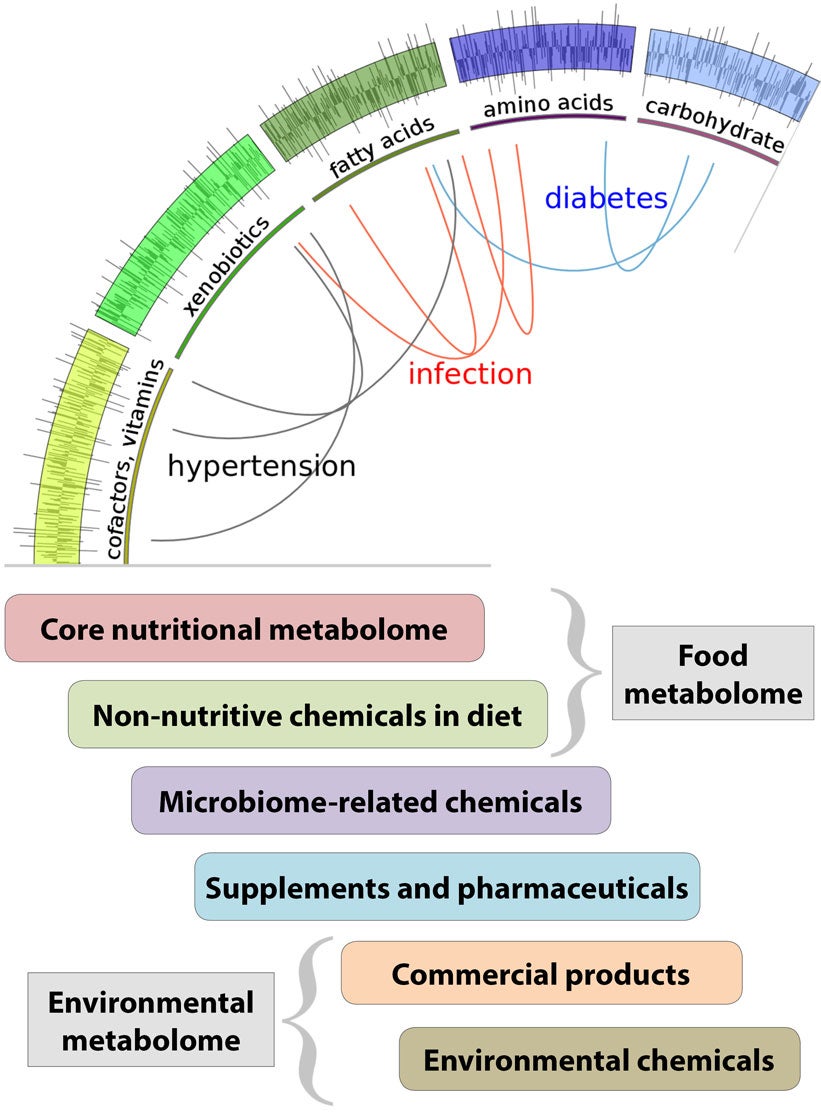
Mapping the exposome is ultimately about understanding the divergence between our genetic predispositions and our biological reality. It requires not just studying environmental chemicals—it is about studying all of the chemicals, endogenous and exogenous alike, that influence human biology. Yet today, a typical laboratory analysis of a blood sample is run through an instrument called a gas chromatograph that groups fractions of the blood by their volatility, and then these fractions are subjected to more detailed analysis by a specific chemical test or by mass spectrometry. In virtually every such analysis, for whatever purpose, there are hundreds of peaks (compounds) that are simply unknown. This is where increased efforts in fundamental research in mass spectrometry and bioinformatics could yield great returns.
What made rapid mapping of the human genome possible was sequencing of many small parts of the genetic code whose function was known—so-called shotgun sequencing—and then putting the pieces together. The equivalent for mapping the exposome is an approach that identifies the environmental mediators that have the greatest effect on human biology and looks for the markers associated with them. As it turns out, many different institutions—from the Centers for Disease Control to university laboratories working under grants from the National Institutes of Health—have collected and stored blood samples from specific individuals whose subsequent medical histories are known at least in part. The Department of Defense, for example, has over 100 million blood samples from soldiers collected at enlistment and again post-deployment from a specific mission such as Iraq. And these archival blood samples turn out to be a treasure trove for linking specific impacts to biochemical markers and thus building up the map of the exposome.
One recent example of this process at work is a child health study conducted by the University of California at Berkeley with blood samples going back 50 years from families that lived in California’s central valley—where concern over pesticide exposure from the intensive farming activity has long been a concern. The Berkeley study looked at the blood from the mothers of daughters who subsequently (as adults) were diagnosed with breast cancer, and sure enough, found clear evidence of pesticide residues that would have exposed those daughters in utero. The study is now extending the analysis to the granddaughters.
Another study at Emory University looked at glaucoma, a condition in the eye that leads to nerve damage and blindness and whose exact cause is not known. The study compared biomarkers in blood samples from people suffering from glaucoma with comparable individuals who did not have glaucoma, and turned up four specific markers in the glaucoma patients. When analyzed, the markers turned out to be related to steroids produced only by fungal infections. Further work to see if the fungal infections might be causal is underway. Another study found markers from two specific environmental chemicals associated with macular degeneration, a condition in older adults that damages the retina and is also a common cause of blindness. These results are likely only the forerunners of a flood of new findings. More fundamentally, these are the beginning of a more complete knowledge of the chemistry of nurture, of life as it is really lived amidst a complex and changing array of foods, industrial chemicals, environmental toxins, and infectious agents.

How to Map the Exposome
While genomics serves as a blueprint of a human being, personal health is also a product of life history and interactions with the environment. Big data from high-throughput molecular technologies that detect the chemical markers in blood or other tissue samples—from metabolic processes, from immune system processes, from activated genes, from foreign proteins—thus provide critical information toward health monitoring, disease diagnosis, and treatment. The key technology that enables detection of these molecular footprints of biological processes is ultra high resolution mass spectrometry, which in effect sorts these chemicals by their unique mass. Such machines yield nearly 10 times the data quality of earlier versions, cost about $1 million apiece, and must be supported by laboratories, trained technicians, and biometric specialists.
In a good lab, with a current model ultra high-resolution mass spectrometer, it is possible to separate and identify between 5,000 and 20,000 new biomarkers per year, including building the algorithms to automate future recognition of that marker. Thus with 10 instruments spread across several universities, mapping 100,000 biomarkers a year might be feasible. With sufficient research funding, a comprehensive, annotated database amenable to big data tools and to inter-comparison with rapidly expanding genomic databases might be feasible within five to seven years. Unlike the $1 billion human genome project, the likely price tag to map the exposome is more like $100 million. Nonetheless, the scale and duration of such a project puts it outside the range of typical federal research grants, which in turn discourages many scientists from tackling it. Moreover, research on the environment has historically been within the domain of the National Science Foundation, while human health falls under National Institutes of Health—the exposome doesn’t fit neatly into either funding structure. Hence there is an extraordinary opportunity for private philanthropy. Quite apart from the fundamental knowledge of how genes and environmental factors interact to influence human illness or capacity, the potential of identifying a person’s lifetime exposure history from a sample of blood, quickly and cheaply—like the $1,100 tests for an individual gene map now available—is a tantalizing prospect. Moreover, the technology of mass spectrometry is advancing rapidly, so faster, more automated, higher resolution instruments will be available, and perhaps, with a concentrated effort, even more sophisticated ways of identifying our exposome. And given the potential value of such services, once academic basic research has shown the feasibility and built the core knowledge, it seems likely that the biotech industry would invest additional capital to extend the database and the inferences that can be drawn from it to improve human health and well-being.