Johann Wolfgang von Goethe, the 18th-century poet and philosopher, believed life was hardwired with archetypes, or models, which instructed its development. Yet he was fascinated with how life could, at the same time, be so malleable. One day, while meditating on a leaf, the poet had what you might call a proto-evolutionary thought: Plants were never created “and then locked into the given form” but have instead been given, he later wrote, a “felicitous mobility and plasticity that allows them to grow and adapt themselves to many different conditions in many different places.” A rediscovery of principles of genetic inheritance in the early 20th century showed that organisms could not learn or acquire heritable traits by interacting with their environment, but they did not yet explain how life could undergo such shapeshifting tricks—the plasticity that fascinated Goethe.
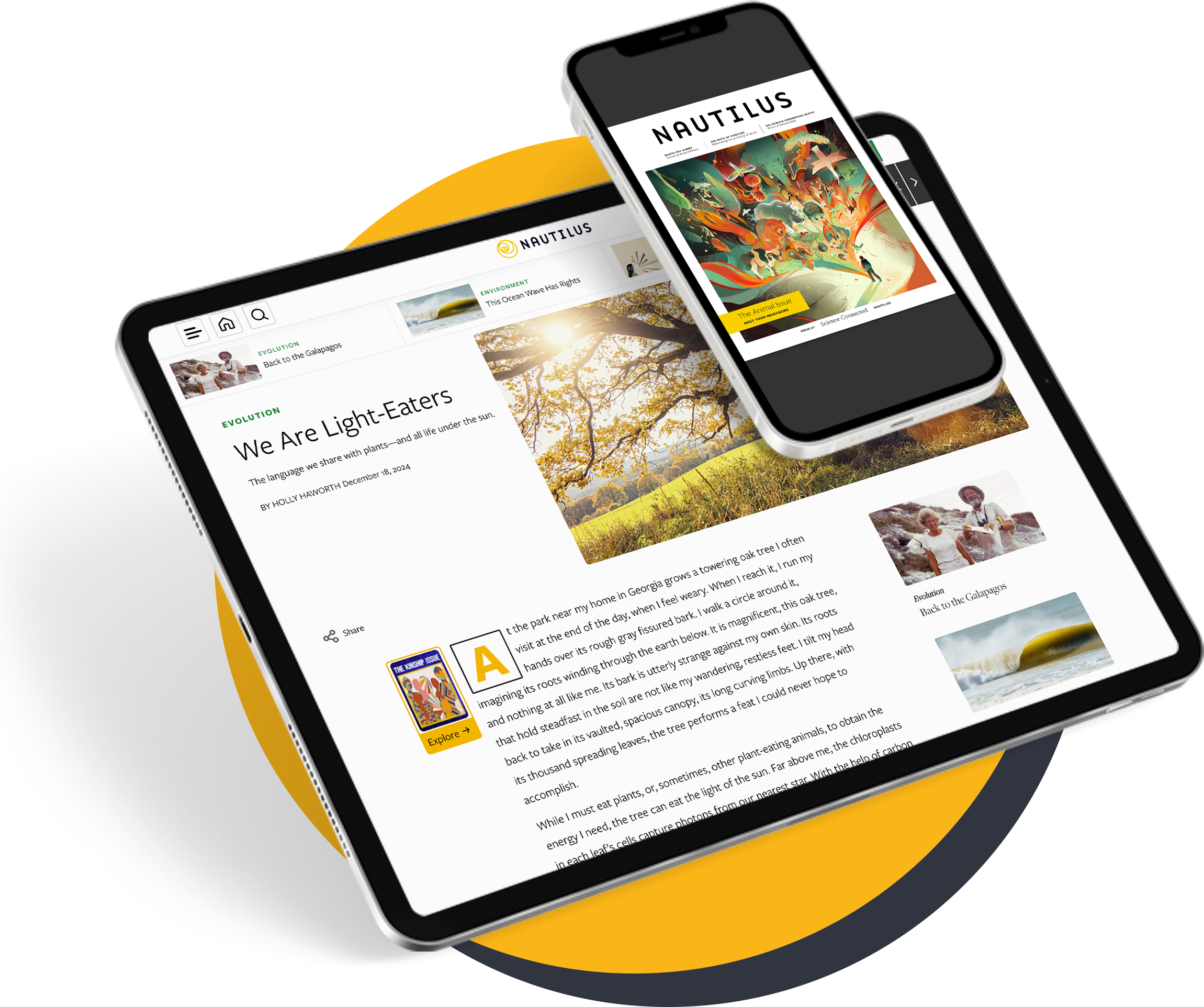
A polymathic and pioneering British biologist proposed such a mechanism for how organisms could adapt to their environment, upending the early field of evolutionary biology. For this, Conrad Hal Waddington became recognized as the last Renaissance biologist. This largely had to do with his idea of an “epigenetic landscape”—a metaphor he coined in 1940 to illustrate a theory for how organisms might regulate which of their genes get expressed in response to environmental cues or pressures, leading them down different developmental pathways. It turned out he was onto something: Just a few years after coining the term, it was found that methyl groups—a small molecule made of carbon and hydrogen—could attach to DNA, or to the proteins that house it, and alter gene expression. Changing how a gene is expressed can have drastic consequences: Every cell in our body has the same genes but looks and functions differently only due to the epigenetics that controls when and how genes get turned on. In 2002, one development biologist wondered whether Waddington’s provocative “ideas are relevant tools for understanding the biological problems of today.”
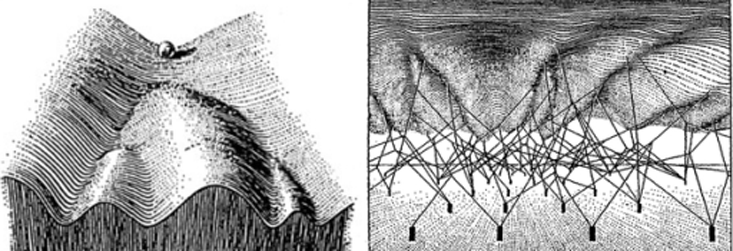
In fact, they are. Fifteen years on, a team of Johns Hopkins researchers, inspired by Waddington’s “epigenetic landscape,” recently came up with a powerful new way of seeing epigenetics. Andrew Feinberg, a member of the research team and the director of the Center for Epigenetics at John Hopkins School of Medicine, said that the team’s results “could have major implications for how we treat cancer and other aging-related diseases.” Their approach, detailed in a Nature study, enlisted information theory, a study of the storage and communication of information. By calculating the propensity of DNA methylation to change across regions of the human genome, they could, in effect, understand cells’ methylation landscape as a communication system.
Nature is more complex than we previously thought.
DNA methylation changes throughout a cell’s lifespan. Consider that stem cells are plastic, highly adaptive to changing their signaling patterns (which genes are expressed) but soon turn into differentiated adult cells that have faithful signaling patterns and convey information in reliable ways. Even adult cells have some degree of stochastic variation, or random fluctuation. But this variation is not just noise, but a cell showing it is still young and adaptable—it allows immune cells to rapidly change to respond to environmental insults, brain cells to form new connections, or cells to turn off genes so that they can repair them. Aging cells begin to teeter into states of uncertainty in which they can no longer reliably conserve DNA methylation information. Since “quantifying uncertainty forms the basis for information theory, we thought that applying information theory to epigenetic data was a natural thing to do,” said Feinberg, who with Hopkins colleague John Goutsias, was a senior author of the paper.
They analyzed DNA methylation across the entire genome of 35 types of cells. This created a big picture of how changes occur in DNA methylation across the genomes of cell types as those cells develop from stem cells into more defined lineages. The researchers weren’t surprised to see that the methylation patterns of cancer cells were computationally distant from healthy adult cells. They were surprised to see that cancer cells were even farther computationally from stem cells, a finding that challenges the conventional thinking that cancer cells revert to a molecular state that is similar to stem cells.
“That was indeed a shocking finding for us,” Garret Jenkinson, the paper’s lead author, and today an instructor of biomedical informatics at the Mayo Clinic, said. Cancer cells are less like stem cells than we thought, and “that just does not fit with the current state of understanding. It indicates that nature is more complex than we previously thought.”
In a 1948 paper, “A Mathematical Theory of Communication,” Claude Shannon, the founder of information theory, described information as a set of messages sent over a noisy channel that could be measured in “bits,” a series of 1s and 0s. A signal could be sharpened, such as shouting down a pipe, or washed out with noise, an effect of other signals that can muddle the clarity of a signal being communicated.
“What we realized,” Jenkinson said, “is that DNA methylation” can be conceived as a bit, a 1 or 0, because when it comes to methylation there are only two options: unmethylated or methylated. Specifically, methyl groups often bind to a particular part of DNA—what’s called a “CpG dinucleotide,” a cytosine (C) paired with a guanine (G) on the phosphate backbone of the double-helix. These dinucleotides often occur in clusters, called “CpG islands,” which are frequently flanked by less dense regions, called “CpG shores.” Thus, DNA methylation is binary information that is either conserved, or not conserved, through time and cell division.
“At a single CpG site you have a binary signal (methylated or not) that is heritable (transmitted from parent cell to its progeny),” Jenkinson said. “This transmission is imperfect (the bit can be flipped), and mathematically can be described as a binary asymmetric channel,” meaning the CpG dinucleotide tends to gravitate to a DNA methylation state. The Hopkins team defines this gravitation as an “energy potential,” a tendency for a methylation pattern to remain or change. A genetic region analyzed over hundreds of bases could be either unmethylated or fully methylated or any pattern in between. A low potential is a methylation state that is easy to change, while a high potential is a state that is hard to overcome. This methylation status is determined by the density of CpG nucleotides in the neighborhood and the activity of enzymes which add or strip methyl groups—for adult cells this information is tightly regulated.
The cellular machinery that regulates this epigenetic code is a rapidly developing area of research for scientists who want to learn to modify the code as a possible therapeutic mechanism. For instance, it has long been known that enzymes add methylation to DNA, but it was also long believed that methyl groups eventually fall off of DNA, like shingles breaking off a roof. Only in the last couple years has it been discovered that proteins exist that actively strip methyl groups from genetic regions.
DNA methylation is binary information that is either conserved, or not conserved, through time and cell division.
How responsive and sensitive a gene is to the machinery regulating its DNA methylation—so called “entropic sensitivity”—is critical for a cell’s function. Stem cells may be highly responsive to this machinery, and hence very “plastic,” while the loss of sensitivity to this machinery, and thus the gene’s increasing rigidity, seem to be hallmarks of aging and cancer. Adult cells such as intestinal cells, or liver cells, need to maintain their responsiveness to this machinery and maintain their epigenetic memory of which genes to turn on, a task that depends on its ability to listen and respond to machinery that maintains it. But aging cells are less responsive to machinery that regulates their methylation status, and are more rigid, often having long blocks of methylated or un-methylated genetic regions. These long stretches of the genome can have a lot of entropy, meaning they can change at any time, quite independently from the machinery that normally regulates their methylation. As a result, genes may be far less adaptive to turn on or off as needed in response to various environmental stimuli (as genes are needed to do, as immune cells spring into action, neurons rewire, or as cells repair and fix themselves) but these long stretches of the poorly regulated genes may also be more susceptible to double-strand breaks and other forms of catastrophic damage that can lead to cancer.
As a motivating example, the authors looked at methylation over a short region in the WNT1 gene, which builds a signaling protein key to cell fate decisions, meaning the commitment a cell makes to become a specific differentiated form, such as an intestinal cell. Colorectal cancers occur in the intestine and are among the top three most common cancers in both men and women. In a healthy colon, this gene has little methylation and exhibits high energy potential, meaning it tends to gravitate to an unmethylated state—it is highly regulated. This implies “that significant energy is required to leave the fully unmethylated state,” Jenkinson said. “Any deviation from this state, meaning addition of methyl groups, will be rapidly ‘funneled’ back,” which results in the maintenance of this gene’s low methylation state in a healthy colon. However, in a cancerous colon, WNT1’s methylation states exhibit low potential, meaning deviations from the unmethylated state in WNT1 will be “frequent and long-lasting, leading to uncertainty in methylation status.” It is more entropic, and the information transmitted by this gene is no longer regulated—it is information lost.
A Journal of Clinical Oncology paper noted that one of the first recognized alterations in colorectal cancers is widespread loss of DNA methylation, which relates to a cell’s age and can lead to the worst form of damage—a double-strand DNA break. DNA methylation changes are so elementary to cancer cells they are now being pursued as biomarkers; in other words, molecular signatures predictive of cancer. But it is so far unclear whether researchers will be able to reliably alter epigenetic code, or more importantly, to restore the normal plasticity of cells to turn back the clock on aging.
Jim Kozubek is the author of Modern Prometheus: Editing the Human Genome with Crispr-Cas9.
Lead image credit: kotoffei / Shutterstock.