Reprinted with permission from Quanta Magazine’s Abstractions blog.

An overarching theme in the story of evolution, at least over the past half billion years or so, is rising complexity. There are other themes, of course, but life has undoubtedly become more complicated since its origin. Early cells globbed together to form multicellular coalitions. Those developed more complex bodies and lifestyles as the millennia passed, finding ever more varied ways to make a living. You might expect that as bodies became more complex, genomes did as well.
But a recent study appearing in Nature Ecology & Evolution shows that not to be the case—at least for jellyfish, humble organisms that evolved at a crucial juncture in animal history. They did not need more genes—or even notably different ones—to power their giant leap in complexity. This new study adds to a growing body of work that casts doubt on finding straightforward genomic signatures of the evolution of complexity.
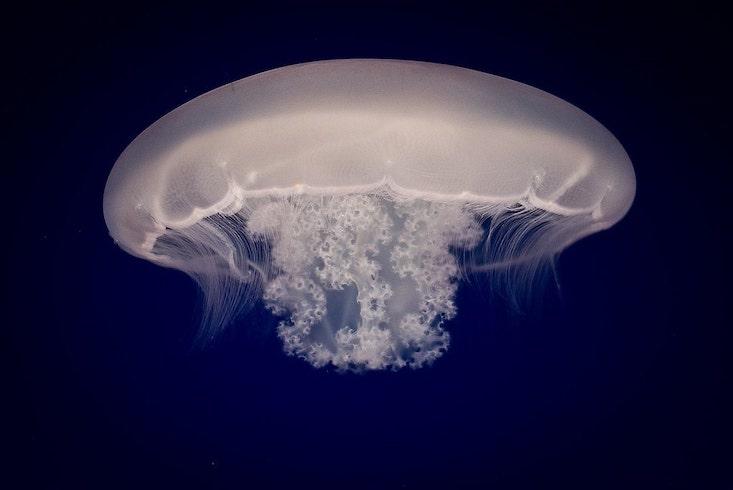
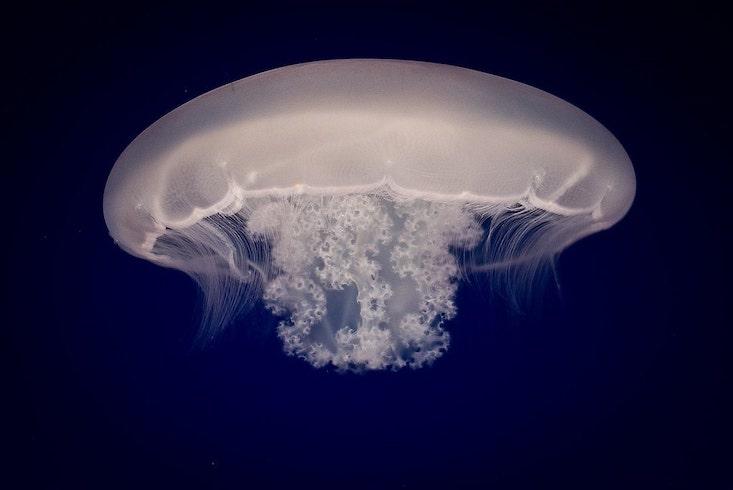
Jellyfish sit alongside sea anemones, coral and hydra on the Cnidarian branch of the animal tree of life. The fork leading to cnidarians represents the final turnoff before animals become bilaterally symmetric, which makes them an interesting group to study because of the greater complexity that came with that later innovation. Jellyfish begin their lives much like their cousins, as sessile polyps anchored to the seafloor, scrounging for food in the passing currents. Unlike their cousins, they eventually break free and transform into a free-swimming form known as a medusa, “what we normally think of when we think of jellyfish,” said David Gold, a biologist at the University of California, Davis, who led the study.
Gold explained that the medusa stage represents a quantum leap in complexity. Medusas actively hunt plankton and navigate the water column with neural sensory structures that detect light and orientation. To go from being a stationary polyp to a floating medusa is almost akin to humans evolving the ability to swim through the air and capture birds with springy, netlike appendages.
The danger when gauging animal complexity, however, is that it’s hard to be objective about it. Mansi Srivastava, a biologist at Harvard University who studies animal complexity and was not involved in this study, cautions that how we view complexity can have more to do with us than with what we’re trying to define. Because we’re so different from sponges and jellyfish, we can miss complex metabolic pathways or other features of “simpler” animal life, leading us to conclude simplicity where there is nuance.
Still, Srivastava and Gold agree that if you tie complexity to life history, jellyfish are more complex than their cnidarian kin. But how they made this jump was unclear. “We just had no idea of what sort of genetic changes were needed to go from this more simple lifestyle to this more complex lifestyle,” said Gold. To find out, the researchers decided to sequence the genome of Aurelia, the moon jellyfish, and then compare it to those of cnidarians without medusas.
If a radical shift in life history requires a big boost in gene content, the Aurelia genome should be riddled with novel genes unique to jellyfish. Instead, Gold found that, broadly speaking, “there really isn’t a whole lot of difference between Aurelia and their relatives with simpler lifestyles.” There were some new genes, but no more than you might expect from any distinct group.
This finding was not terribly surprising to Gold because the genomes of other, much more disparate species also look fairly similar if you squint at them. Gold already had a more nuanced hypothesis: When it comes to building a body, it’s not just what genes exist that matters but also when they’re used. If you’re looking for genes that build complexity in jellyfish, then it makes sense to look for them when that complexity is manifesting, during medusa development. Gold thought that genes unique to jellyfish would be active during the transformation from polyp to medusa.
But to his surprise, that’s not what he found. New genes unique to jellyfish were no more likely to be expressed in the medusa stage, or any stage of development, than other, older genes were. “At the broad genetic level, it doesn’t seem like you need major changes in the genome to make these big changes in your life history,” Gold said.
An alternative explanation accounts for the lack of differences by flipping around the origin story. Perhaps complexity, in the form of a medusa stage, existed in early cnidarians but was lost in all related groups living today except jellyfish. Either scenario would produce a similar genomic signature, although Gold thinks the latter less likely. Comparing more cnidarian sequences at different stages of divergence will eventually reveal the answer.
“It’s not that surprising that the jellyfish didn’t just invent a whole bunch of new genes to make a medusa stage,” said Srivastava, “but we don’t know until we look.” She was intrigued by the finding that novel genes weren’t overrepresented in the medusa stage because it suggests that “very different body plans can arise by connecting the same genes in different ways.”
Gold’s results broadly align with those from another jellyfish genome, Clytia. That research, too, found no large role for novel genes. To add to the mystery, there were even hints that in Clytia, more ancient and conserved pathways played a larger role in medusa development.
In any case, for now, the genetic changes that orchestrate this metamorphosis in jellyfish remain unknown. The transformation may depend on regions of the genome that don’t encode proteins, but instead regulate when genes are turned on and off. Perhaps it’s easier for life to innovate by rearranging its existing gene networks instead of evolving scores of new genes. Or perhaps the broad first pass at the genomes simply missed a handful of coding genes that play an outsized role in the process.
The Aurelia genome joins a growing number of studies that complicate our view of complexity. When scientists began comparing the genes and genomes of different branches of the tree of life, they expected to find vast differences, but instead discovered remarkable similarity. For example, humans and cats share about 90 percent of our genes; we share nearly two-thirds of our genome with fruit flies, despite being separated for approximately 800 million years.
Even the earliest animal lineages harbor unexpected complexities. When Srivastava and colleagues sequenced the first sponge genome in 2010, they were stunned to find genes that built the brains and muscles of other animals already present in this brainless, muscleless sponge. “The genes are the same, but clearly they aren’t working together to do the same things,” she said.
As more genomes get sequenced, researchers will be able to move beyond broad brush descriptions. “I think genome sequencing is a really great tool for generating hypotheses,” said Srivastava. “And then the sequence itself provides us with a tool to more effectively test those hypotheses.” Armed with sequence data and functional tools like CRISPR, scientists can start tweaking individual genes and gene networks to disentangle the relationship between their connections and the varied forms they build.
“I can give you a list of genes that seem to be associated with an increase in complexity,” Srivastava said, “but at some level that’s not very powerful information. It can’t explain why a human looks like a human and a sponge looks like a sponge.” According to her, “the next big advances will come from sort of the hard work of doing experimental biology,” and she’s excited to see where that hard work takes us.
Johnathan Lambert is an evolutionary biologist turned science writer, currently working as Quanta Magazine‘s writing intern. Before that, he was a AAAS Mass Media fellow at the Dallas Morning News, and was also an assistant producer on the science podcast for kids from American Public Media, Brains On!. He is currently a Ph.D student at Cornell University.