Every year the paleontologist Alan Cooper meets up with a band of miners in the Yukon. As the miners go about their work, spraying massive jets of water at frozen mud and silt to excavate gold, they also expose animal remains from the Pleistocene, between 2 million and 12,000 years ago. Back then, North America was a riot of big mammals. Antelope, camels, llamas, and indigenous horses roamed the vast plains; lions, saber-toothed cats, and dire wolves prowled for prey; and giant ground sloths lumbered along, stripping tree branches of leaves and fruit.
But, by the time the planet transitioned to the current geological epoch, the Holocene, the vast majority of those species had gone extinct, most likely due to climate change or hunting by humans. Climate change in the Pleistocene was “huge, frequent, and rapid,” says Cooper, director of the Australian Centre for Ancient DNA at the University of Adelaide. “Sometimes a change of 10 degrees centigrade over a space of a decade or two.” It’s difficult for animals to cope with such dramatic shifts through standard evolution by natural selection, which often takes decades—even millennia—to spread advantageous genetic mutations and hone adaptations.
Damage inherent to ancient DNA could open a window onto ancient epigenetics.
Yet, somehow, a few megafauna survived the mercurial Pleistocene, including a few species of bison. Their living descendants, the North American wood and plains bison, are the largest land animals on the continent today. A great puzzle for paleontology is what made the bison so resilient. How was it able to adapt when so many others failed?
Cooper thinks part of the answer lies beneath the Yukon permafrost. With the right techniques, he can pull a secret history from the DNA in the fossils he collects. Their genetic material conceals a dimension of evolution that scientists are just starting to understand, known as epigenetics. Epigenetic mutations are potentially heritable, but, in contrast to genetic mutations, they do not alter DNA sequences. Instead, they manifest themselves in patterns of molecules glued to DNA that help determine which genes are active. The epigenome, as these patterns are known, is highly malleable, continuously adapting to an organism’s physiology, as well as to fluctuations in its external environment. Some of these epigenetic adaptations become relatively stable.

If they consistently travel from one generation to the next in animals and people—which researchers still debate—they represent a neglected form of evolution, distinct yet parallel to typical genetic mutation. And even if they do not, widespread epigenetic changes within each generation could help groups of animals to swiftly adapt to environmental stress, such as big fluctuations in climate, dwindling food sources, or, in the case of people, prolonged warfare.
For the moment, though, the importance of epigenetics for long-term evolution is an open question. To help answer it, Cooper and other scientists in the young field of paleoepigenetics are extracting DNA from fossils and comparing epigenomes across great swaths of time. If a genome is like the primary historical account of how a species evolved—the one that most scholars reference—the epigenome is a palimpsest of crucial annotations and revisions that is still being discovered. Ancient epigenetics could help explain how the environment sculpts a species over time, why some species outlive others, and how species form in the first place. It won’t be easy, though. For one thing, it’s going to require an awful lot of digging.
Although researchers have been recovering DNA from the remains of primeval creatures since the 1980s, resurrecting ancient epigenomes from fossils was not possible until about six years ago. Like the genome, the epigenome resides in a cell’s nucleus, its genetic command center. Teams of enzymes zip about gluing and detaching various molecules to and from strands of DNA. You can think of these molecules as DNA’s removable padlocks and welcome mats: They either block other cellular machines from accessing certain sequences, effectively silencing those genes, or encourage them. When looking for traces of these epigenetic markers in fossil DNA, researchers focus on the most durable one: methyl groups, which are usually tagged onto the nucleotide cytosine (the “C” in DNA’s four-letter alphabet, GATC). In general, methylation silences genes.
When DNA has been sitting around for tens of thousands of years, it inevitably degrades. Cytosine often degrades into uracil, a nucleotide usually found in RNA. DNA sequencing machines misread these damage-produced uracils as thymines (“T”), so researchers often eliminate them with various chemical treatments. In 2009, Adrian Briggs was trying to do just this with Neanderthal DNA. At the time, he was a graduate student at the Max Planck Institute for Evolutionary Anthropology in Germany, studying under Svante Pääbo, the world’s leading expert on ancient DNA. Despite Briggs’ best efforts, the Neanderthal DNA sequences still contained an unexpectedly high number of thymines.
A growing body of evidence says trauma, famine, and violence in living human populations have long-lasting epigenetic effects.
Through conversations with colleagues, Briggs learned that only unmethylated cytosines degrade into uracil, whereas methylated cytosines decay into thymine. That was the likely source of the extra Ts. Briggs realized that he could turn this seeming nuisance into an advantage. First, he would get rid of all the uracils in his sample of Neanderthal DNA. Then he would compare the Neanderthal genome to the modern human genome, looking for locations where modern humans have a C and Neanderthals have a T. Some of those positions might represent a genuine mutation, but most would indicate a methylated Neanderthal cytosine that had degraded to thymine. Damage inherent to ancient DNA could open a window onto ancient epigenetics.
In 2010, Briggs, Pääbo, and their colleagues reported that they could reconstruct methylation patterns in DNA from the bones of a 40,000-year-old Neanderthal and wooly mammoth. It was an unprecedented achievement. Two years later, Cooper and his team used a somewhat different and more precise technique to analyze methylation patterns in DNA from a 26,000-year-old Steppe bison fossil from the Yukon. Before sequencing, they used sodium bisulfite to convert all the unmethylated cytosines into uracils, leaving only undamaged methylated cytosines for the comparison. Regions known to be consistently and highly methylated in modern cattle were also highly methylated in ancient bison, suggesting that their reconstruction was accurate.

Even more tantalizing findings have emerged from human remains. Last year, Ludovic Orlando and his colleagues at the University of Copenhagen deduced the methylation patterns preserved in DNA from a 4,000-year-old Greenlander. Not only did the sites of methylation match those in DNA from the hair of living humans, they also made biological sense, permitting the expression of genes crucial for forming the proteins in hair.
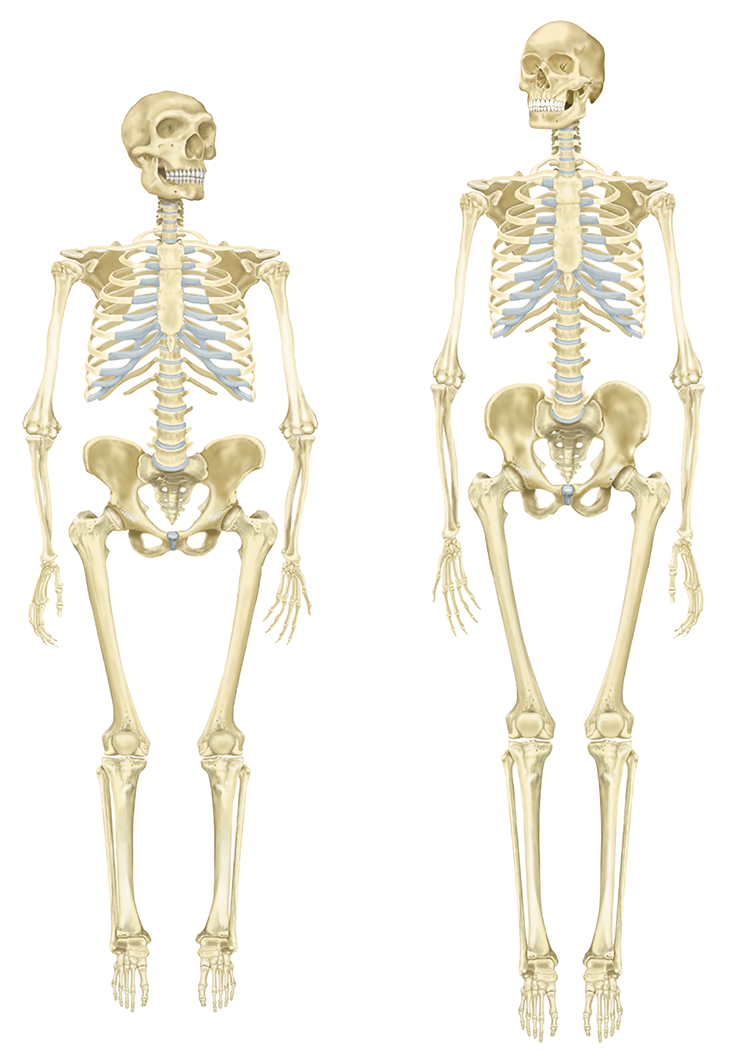
Similarly, Liran Carmel and his team at The Hebrew University of Jerusalem found that most of the methyl groups in Neanderthal and Denisovan DNA synced up with those in the modern human genome, as would be expected for such closely related hominins. In some areas, however, there were clear differences, like with the HOXD10 and HOXD9 genes. Although these gene sequences themselves are identical in humans and Neanderthals, they are more methylated in the latter. They are also crucial for limb development, suggesting that epigenetics might be partly responsible for the evolution of gracile arms and legs in modern humans, as opposed to the somewhat shorter and stockier limbs of Neanderthals. Some researchers have proposed that Neanderthal anatomy was less energy efficient than our own, possibly giving us the advantage when hunting and migrating long distances to escape unfavorable conditions, such as drought or harsh winters.
Most recently, Rick Smith of The University of Texas, Austin investigated epigenetics in Native American remains between 230 and 4,500 years old. Using bisulfite sequencing, he was able to identify methylation signatures in 29 of 30 samples—an unprecedented success rate. He is particularly interested in whether violence, starvation, and other kinds of stress in the last few thousand years left epigenetic impressions in the DNA of Native American tribes. “A growing body of evidence says trauma, famine, and violence in living human populations have long-lasting epigenetic effects, especially if they are experienced early in life. We can look for similar epigenetic patterns in ancient DNA from societies where we have good records showing they went through similar stress.”
Although many researchers are thrilled by the potential of paleoepigenetics, they are also quick to bring up a long list of important caveats. “We are in the birth of this field, and there’s so many potential directions we could go, a lot of questions we can try and answer now. I’m really excited,” says Smith. “But we should proceed with caution and be really careful about the inferences we make.”
Some of the challenges are technical: It is quite rare to recover enough DNA from truly ancient fossils to perform any kind of meaningful analysis. In their first study, for example, Cooper and his colleagues could only extract sufficient DNA from one of six bison skulls. The analysis itself is even trickier: “Almost anything you do affects your DNA methylation pattern,” says Carmel, “but it’s still not entirely clear how to interpret those patterns in all situations.” Methylation signatures can vary immensely from one type of tissue to another, so the epigenetics of teeth, bone, or hair—the kind of tissues that survive the eons—might not be an accurate portrait of an organism as a whole. And different individuals of different ages within a single population may also have markedly different epigenetics, so drawing broad conclusions about an entire species based on one or two samples is tentative at best.
So far, researchers have tried to circumvent these difficulties by focusing on regions of the genome that are definitively known to be strongly or weakly methylated across a given species. In order to make confident conclusions about more variably methylated sections of the genome—the ones where they would expect to find signs of adaptation—they will have to patiently collect numerous specimens. “The only way to tackle this is to work on really huge numbers, hundreds of individuals, because that way you kind of average the biological factors that might be problematic,” explains Orlando. “Then we are not just comparing individuals, but whole sets.”
If epigenetic mutations themselves are inherited in animals and people, and survive for many generations, then they undoubtedly alter long-term evolution.
The even bigger dilemma is the ongoing debate about whether epigenetic changes are inherited, and how often they survive long enough to make a difference in evolution. Scientists have documented numerous cases of epigenetic inheritance in bacteria, fungi, and plants, but similar examples among animals are rarer and usually confined to laboratory experiments. A few long-term studies of human health, most famously one that tracked families during and after the Dutch famine of 1944, suggest that a pregnant mother epigenetically transmits a susceptibility to poor health to both her children and grandchildren, but this effect seems to disappear in subsequent generations. Further complicating things, whereas the biological mechanisms underlying transgenerational epigenetics in microorganisms and plants are fairly well-established, they remain mysterious in animals.
Given these dilemmas and knowledge gaps, some geneticists are highly skeptical about the importance of epigenetics for long-term evolution. A review of epigenetics research published in Cell last year concluded that “environmentally induced epigenetic changes are rarely transgenerationally inherited, let alone adaptive, even in plants. Thus, although much attention has been drawn to the potential implications of transgenerational inheritance for human health, so far there is little support.”
If epigenetic responses to stress are not inherited, then any intriguing discrepancies researchers spot in fossil DNA will necessarily be confined to particular individuals and single generations. Even without a legacy, however, epigenetic mutations could play a subtle and indirect role in evolution: If they give a creature a reproductive advantage within its own lifetime, then they contribute to the spread of that creature’s genes. Some scientists have also proposed that environmentally-induced adaptations sometimes become genetically encoded: Perhaps, through unknown mechanisms, genetic mutations arise that increase the likelihood of establishing certain beneficial epigenetic patterns.
If epigenetic mutations themselves are inherited in animals and people, and survive for many generations, then they undoubtedly alter long-term evolution. The discrepancies Carmel discovered between the Neanderthal and human epigenomes hint at this possibility in our own evolutionary history, but these findings are based on just a few DNA samples. Until researchers amass sufficiently large libraries of ancient DNA, they cannot make more definitive conclusions. Which is why, every year, Cooper and his colleagues return to North America, roaming frigid black slopes and descending into caves in search of promising fossils. So far they have collected several hundred specimens spanning about 50,000 years.
Cooper’s plan is to compare the epigenetics of bison that lived before and after specific periods of stress and see if he can tie any epigenetic mutations to anatomical adaptations that might have helped the animals survive. “It really is a hell of a challenge,” he says, “but it looks very promising at the moment.”
Ferris Jabr is a writer based in Portland. He has written for The New York Times, The New Yorker, Scientific American, Wired, New Scientist, Popular Mechanics, NOVA Next, and The Awl.