There’s a silent epidemic slowly brewing around the globe. It hasn’t claimed as many lives as COVID-19, but if it gets out of control, it will likely prove even deadlier. It’s caused not by a single virus, but by a number of different bacterial species that have been slowly evolving defenses against our once potent antibiotics. Think of it as a sea slowly swelling with superbugs.
Currently, someone in America dies from an antibiotic-resistant bacteria or fungi every 15 minutes, adding up to 35,000 annual deaths. Globally, the United Nations reports that a mind-boggling 700,000 people a year die from infections antibiotics can no longer fight. The report warns that this number could reach 10 million by 2050 if we don’t find alternative therapies against our bacterial enemies.
How did we arrive at this existential threat to humanity? Whether we like it or not, that’s evolution in progress. In the 20th century, we invented powerful antibiotics that destroyed bacterial cells before they destroyed us. In response, the bugs evolved too. They learned to produce chemicals that deflect our antibiotics, so they can continue multiplying. We thought we could outpace bacterial evolution with our pharmaceutical prowess, but the germs beat us, bolstering their defenses. We can’t fight evolution.
Or can we?
David McCandlish, who studies evolution and population genetics at Cold Spring Harbor Laboratory, thinks we can predict it. A computational biologist, McCandlish uses mathematical and statistical techniques to study living organisms, with a goal of building predictive models of how they may evolve. McCandlish believes that using the knowledge of genetic and biological mechanisms of how bacteria develop resistance can help us foretell what they might do in the future, in response to our meds. “If there’s a microorganism that we have a drug against, an antibiotic, or anti-microbial, we want to predict the specific sequence of mutations by which it will evolve antibiotic resistance,” McCandlish says. “So yes, we want to be able to predict evolution.”
McCandlish’s approach draws on the concept of the so-called adaptive landscapes—an idea that looks at organisms’ evolution as peaks and valleys. The adaptive landscapes model views evolution as a hill-climbing process in which the populations with winning genetic traits—or highest genetic fitness—would rise up to the peaks, while those with inferior genetics would remain in the valleys between the peaks. Pictorially, these adaptive landscapes, sometimes called fitness landscapes, look like mountain ranges. In these charts, the horizontal axis depicts the organism’s one or several genes and the vertical one depicts that organism’s genetic fitness. The better-fit populations that were lucky to get “the good” mutations climb to peaks. Those that got “the bad” mutations that weigh them down end up in the valleys.
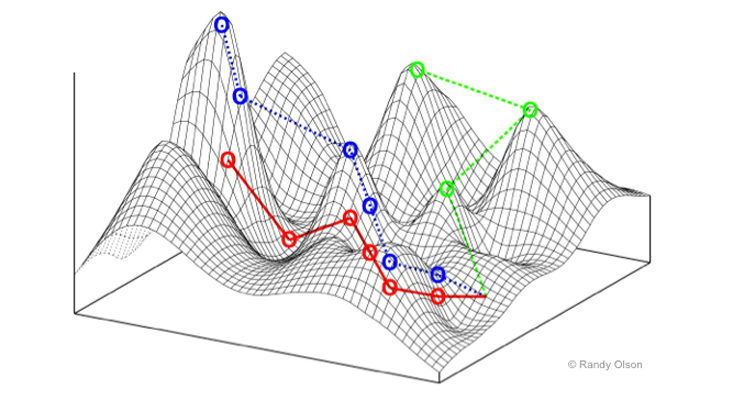
When it comes to bacterial species that are pathogenic to humans, those that climb to the peaks are bad news for us, particularly if that ascent is due to their drug-resistance abilities. If one were to depict some of the deadliest superbugs using adaptive landscapes, the much-dreaded Methicillin-resistant Staphylococcus aureus or MRSA, which claims close to 20,000 lives a year in the United States, would likely be on one of the peaks. So would be the multidrug-resistant tuberculosis, which kills 230,000 worldwide. We’d much rather prefer to see them languishing in the valleys.
How can adaptive landscapes help us predict how the next potential superbug may evolve? To answer that question, scientists first need to understand the cellular, molecular, and genetic forces that drive antibiotic resistance in bacteria. They must also understand how we arrived at this now-looming post-antibiotic era, which, on the grand evolutionary scale, was here in a hurry.
When Alexander Fleming first discovered penicillin in 1928, which led to the creation of the first antibiotic that became widely used in the 1940s and after, humans got an evolutionary advantage over bacteria. We could easily and efficiently kill many bacterial species before they wreaked havoc on our organs, tissues, and bodies. But in the years that followed, we might’ve been a little too overzealous with our antibacterial armor and we used it too much. Doctors prescribed antibiotics to patients when they weren’t necessary. Cleaning product manufacturers put antimicrobial compounds into soaps and dishwashing liquid. Industrial farmers mixed antibiotics into the animals’ fodder, not only because it helped fight infections but also because it made them fatter, which was viewed as a positive side effect.
Consequently, a certain amount of antibiotics washed out or escaped into sewage, trickling into water and soil, where vast populations of bacteria lived. As a result, millions of them died, except for some mutants who survived because they were lucky to have a slightly different assortment of genes that enabled them to develop special cellular defenses. Because the less fortunate bacteria were dead, these lucky mutants went on growing, replicating and passing on the antibiotic-resistant genes.
We thought we could outpace bacterial evolution with our pharmaceutical prowess. But the germs beat us.
“That’s what happens when you put a stressor on a bacterial population, such as an antibiotic,” says Parjit Kaur, professor of molecular genetics and biochemistry at Georgia State University, whose recent paper examines microbial drug resistance mechanisms. “There may be a random mutant that can now withstand that antibiotic, or there can be a spontaneous mutation. And then the resistant bacteria became the dominant population.”
Antibiotics are essentially poisons for bacteria. Once antibiotic molecules enter bacterial cells, they either damage or break the cell, or interfere with the cell’s replication. In either case, the bacterial population eventually dies out. To survive, the bugs must either eject the antibiotic molecules or demolish them, before they destroy the cell.
Different bacteria species fight antibiotics differently, explains Kaur. Some mutate their genes to produce specific efflux proteins that “escort” the trespassing antibiotic molecules out as soon as they enter the cell. That may sound like a sophisticated defense, but it’s not a novel concept. Organisms rely on the so-called transport proteins to bring substances in and out of cells. Typically, these transfer proteins usher in nutrients and spew out waste substances. Some germs simply evolved their genes to generate new transport proteins that ferret out the poisonous antibiotic molecules. “Antibiotics are toxic substances and these specialized proteins pump them right out,” Kaur says. And they go on living and replicating.
Other types of bacteria have double cell membranes—an inner and an outer layer. The space in between the two is called the periplasm, and that’s where these germs stage their antibiotic defense. These bacteria produce antibiotic-cleaving enzymes that work as molecular scissors, which are released into and kept inside the periplasmic space. Once antibiotics enter, these enzymes shred them into a heap of molecular debris. The enzyme mechanism isn’t superbly novel either. Bacteria use enzymes as part of their cellular “housekeeping” to cut other substances, Kaur explains, so it’s possible that some housekeeping enzymes were recruited and developed into “antibiotic warriors.”
Bacteria can also acquire antibiotic resistance genes from each other, and they have multiple ways of doing it. Under certain conditions, some can pick up these genes from the dead bacteria whose DNA had been dumped in the environment. Others can acquire such genes from a virus that penetrates microbial cells but doesn’t kill them—instead it might bring along some genetic material beneficial for bacteria. Lastly, the bugs can “trade” their genes. Two bacteria can saddle next to each other, open their cell walls and exchange their genetic material. Think of it as MRSA snuggling next to an ordinary Staph, whispering. “Psst! I finally figured out how to kick this annoying methicillin bomb. Come close and I’ll share my secret.” And then they both can climb to the peaks.
Modern genetic tools offer scientists the ability to experiment with cells’ genetic mutations and see what evolutionary advantages or drawbacks they may produce.
“You take a bug and put in a bunch of genetic variants and see how fast they will grow,” explains McCandlish. “You take a protein of interest, make lots of different mutations in it, then stick these mutated sequences into cells and try to grow these cells. If that sequence works well, the cells will grow and increase in numbers. And if the sequence doesn’t work, the cells limp along and maybe divide once and then die.”
Scientists are learning to predict the cellular and genetic forces that drive antibiotic resistance in bacteria.
Scientists can identify the growth rates of each of these variants and what effects the mutations had on cells’ growth. They can then determine whether any of these mutations conferred evolutionary advantages. Some mutations may completely wipe out the population while others may imbue it with superpowers. A great advantage of this approach is that it’s versatile—the same concept can be used to predict evolution of any cells—not only microbial, but also tumors.
Scientists are already experimenting with such combos or mutational sequences, as they call them. McCandlish cites one recent study in which researchers inserted a variety of mutational sequences into yeast cells and measured their growth rates. The yeast cells with the most mutations proved to be the sickest, he notes, but it doesn’t have to be so—an occasional combo may prove just as deadly. By noting which mutational sequences give an organism a superb advantage, you can predict which ones will grow and take hold.
McCandlish’s predictive algorithms rely on such studies for data and learning. The more information these algorithms can learn—particularly about pathogenic organisms like bacteria or fungi—the more precise and meaningful their forecasting would be. The yeast was a good playground, but one can do similar experiments for how drug resistance will evolve in bacteria, and see which mutational sequences would enable the bugs to fight antibiotics better.
“Let’s say we have developed a new antibiotic,” McCandlish says. “Now we want to be able to predict how would that enzyme that previously evolved to degrade the last generation of antibiotics, change to degrade new ones? Or maybe it won’t.” Knowing which genes might produce the resistance enzymes or proteins can let pharmaceutical companies stay one step ahead of the bugs.
Having accurate resistance forecasts might ease the pressure of developing new antibiotics. The model might predict that bacteria A can develop resistance to antibiotic X, and later to antibiotic Y, and later antibiotic Z. But the last two rounds of adaptation to Y and Z might weaken the pathogen’s defenses against antibiotic X or disable them altogether. That means a specific germ will now be once again susceptible to antibiotic X, and no new ones would have to be developed to fight it.
Accurate forecasts could allow medics to control or foster the evolution of cells—those of pathogenic microbes or cancer. If we understand how a particular cell evolves on its fitness landscape, we can design drug treatment strategies. “If we know that one specific drug will push bacterial population to evolve resistance by one or the other of these peaks, then maybe by applying the second drug we can choose which way it evolves,” McCandlish says. “Or we can try to push the bugs to evolve to a peak where we know we have a second drug that crushes that peak. It would be like setting a trap.”
While those drug traps may not be available in your neighborhood pharmacist tomorrow, overall prediction models might help us get a leg up in the evolutionary race against microbes to avoid the dreadful 10 million deaths by 2050. Once the models are working, they would restore our evolutionary advantage over pathogens. Superbugs will keep climbing the evolutionary peaks. But we will be there waiting for them—with our antibiotic armor at the ready.
Lina Zeldovich grew up in a family of Russian scientists, listening to bedtime stories about volcanoes, black holes, and intrepid explorers. She has written for The New York Times, Scientific American, Reader’s Digest, and Audubon Magazine, among other publications, and won four awards for covering the science of poop. Her book, The Other Dark Matter: The Science and Business of Turning Waste into Wealth, will be released in October 2021 by Chicago University Press. You can find her at LinaZeldovich.com and @LinaZeldovich.
Support for this article was provided by Cold Spring Harbor Laboratory. Read more on the Nautilus channel, Biology + Beyond.
Lead image: CLUSTERX / Shutterstock