All immune systems are not created equal. The evidence is all-too familiar: We all know that work colleague who never so much as sniffles, even when everyone else in the office is out sick. These days, immune system inequality plays out with especially grim consequences, as one person with COVID-19 dies, while another in a superficially similar state of health experiences only mild symptoms. But why our immune responses vary so widely—and how to make everyone’s system more resilient—is one of the great mysteries in medicine.
The search for answers has led Hannah Meyer to a class of white blood cells known as T-cells, and to their point of origin in the thymus, a bean-shaped organ located near the heart. Meyer, bioinformatics expert and a fellow at Cold Spring Harbor Laboratory, notes that T-cells not only identify and eliminate cells infected with foreign invaders like SARS CoV-2, they also signal other parts of the immune system to mobilize when needed. Those functions depend on the ability of T-cells to reliably distinguish dangerous pathogens from the body’s own cells, a skill that they acquire in the thymus…somehow. “We know the thymus represents everything,” Meyer says, “but we don’t know how it’s regulated.”
Within the thymus, T-cells and epithelial cells interact in virtually limitless combinations, sequences, and distinct micro-environments. It’s possible to observe these interactions directly in a lab, but trying to explore them all empirically is like trying to anticipate the outcome of a chess game by trying out every possible move: overwhelming. Meyer is designing computer simulations that model the combinations virtually and determine which ones are more significant for the training of T-cells in the thymus. Those results can then allow much more tightly targeted laboratory studies.
Why our immune responses vary so widely is one of the great mysteries in medicine.
Lab studies of immune cells and computer models of immune responses have often existed in separate academic realms. Meyer is now integrating the two, as part of a larger movement called computational biology. “We need experimental and computational approaches together to achieve a baseline understanding of what ‘healthy’ means and to understand what might have gone wrong in certain pathologies,” she says.
In the Meyer lab, her team designs software to simulate conditions experienced by T-cells. Researchers can plug in environmental variables, assess the probabilities of various cellular responses, and develop experiments to compare the model’s results with the behavior of actual immune cells. The goal is to figure out what it takes to turn a newborn T-cell into a focused disease fighter—and eventually, to map out the links between infectious disease, cancer, diet, and the immune system.
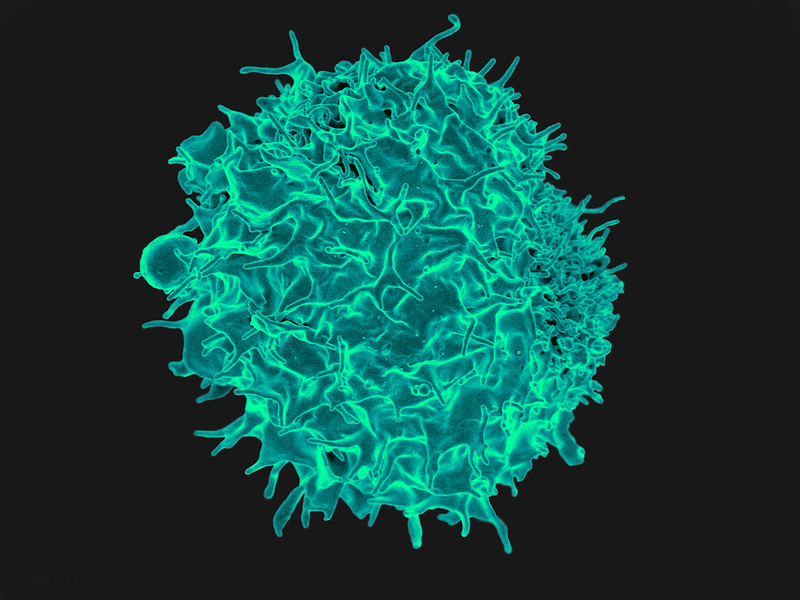
The journey of the T-cells begins during a person’s childhood, when immature T-cells follow a trail of chemical signals that leads them to the thymus. T-cells are born in the bone marrow, initially equipped only with the necessary sensors (called receptors) to detect these chemical bread crumbs. Before they leave the thymus to fight disease, they must develop a whole additional set of specific receptors that allows them to identify every type of healthy cell, tissue, and protein; otherwise, they will end up attacking the wrong target. “T-cells have to learn about anything they could encounter elsewhere in the body, and they have to do it without traveling,” Meyer says. Given that the average body contains 30 trillion cells of some 200 varieties, each T-cell needs an extremely thorough education.
That education comes from specialized cells within the thymus, the epithelial cells. These cells have the ability to present small snippets (called epitopes) of all the proteins a T-cell could encounter throughout the body, thereby teaching T-cells the “look” of virtually any healthy cell and tissue. As a result, when T-cells finally head out of the thymus on their disease-fighting mission, they know that any cell with a foreign, unknown look to it must be sick and dangerous.
Researchers broadly know that thymic epithelial cells train T-cells by enabling them to see all protein epitopes of the healthy self, but they are lacking the specifics. They don’t know how the training is scheduled, how many epithelial cells a T-cell has to visit before it is considered to have passed its education, or whether the T-cells must be exposed to the different epitopes in a specific sequence. Understanding such details might explain some of the key differences between effective and flawed immune responses.
“T-cells have to learn about anything they could encounter in the body, and they have to do it without traveling.”
Two years ago, Meyer was working on a separate but similarly complex problem: What are the differences between a healthy heart and one that is prone to heart diseases? As with her current research, she turned to computer models and simulations to understand the key genetic and physiological components that make a heart beat efficiently. To help bridge the gap between experimental and model studies of the human heart, she developed a suite of computer programs, including the phenotype simulator. The phenotype simulator translates genotype (an organism’s genetic information) into phenotype (the organism’s observable traits). For example, brown hair is a phenotype that results from the genotype for hair pigmentation. The translational outputs can be quite complex, given the tremendous number of environmental factors and feedback mechanisms within the body that can influence phenotype.
Meyer is now applying the computational methods that helped elucidate the genetic components of heart physiology to her favorite puzzle in biology: the education of T-cells by thymic epithelial cells. In this case, her simulations allow researchers to plug in multiple important factors (such as the localization and duration of the education) and then see a range of likely immune-related characteristics that the cells might acquire in response. The resulting datasets provide what Meyer calls a “ground truth” about the ways that immune response differs across these simulated scenarios. By experimenting with different inputs, researchers can probe specific cellular interactions and relationships that affect immunity. Those results can then guide lab studies to search for associated molecular changes in the T-cells.
Meyer focuses specifically on interactions between T-cell and epithelial cells in the thymus, but her simulations are proving far more widely useful: T-cell interactions in different tissues and with different interaction partners are possible. For instance, studies show that autoimmune disease, in which the body attacks its own cells, is correlated with genes and environment, but the cause-and-effect relationship is undetermined. Meyer’s simulation offers a way forward by allowing researchers to isolate individual processes influencing T-cell behavior.
Meyer’s colleague Semir Beyaz, an immunology researcher at Cold Spring Harbor Laboratory, is particularly fascinated by using Meyer’s computational methods to examine the link between cancer, diet, and the immune system. Studies show that high-fat diets contribute to greater tumor formation in mice, apparently by impairing the immune system’s ability to weed out cancerous cells, but scientists don’t know what processes associated with fat lead to weaker T-cells. There are so many variables and mechanisms at play that Beyaz calls it a “multidimensional data mess.”
To cut through the clutter, Beyaz collaborates with Meyer to seek causal relationships between diet and cancer phenotypes that might have taken years to recognize without computer assistance. The computational analysis and simulation generate new hypotheses, which he can then use to devise an experiment and study whether that dietary factor triggered a response directly, or whether it is mitigated through immune cells such as T-cells. “As a bench scientist, you never know if something is actually regulating the process you’re studying until you do functional experiments like turning the process off or over-activating it,” Beyaz says.
Meanwhile, Meyer is building on her previous success in software development by developing ways to help experimental researchers like Beyaz to identify those kinds of processes more effectively. Her new software focuses on individual cellular interactions and “treats each cell as an independent agent,” she says, allowing simulated immune cells to move naturally around a 3D grid and keeping track of their interactions along the way. Meyer wants to find out if different routes, interactions, or sequences of interactions in the thymus alter a T-cell’s immune functionality. The results will again suggest experimental follow-ups. If her models indicate that T-cells traveling on a certain course are more likely to fail and attack healthy cells, for example, researchers could investigate whether there is a preferred order of T-cell migration in the thymus, or if the cellular maturation process depends on location.
Genetics and immunology researchers around the world are already relying on Meyer’s software for their work; her software suite for genetics analysis, including phenotype simulator, is freely available and has been downloaded more than 34,000 times. This established user group reports problems, which then helps Meyer develop revisions to fine-tune the simulator for specific research tasks. “Making other people’s work easier is a big highlight,” she says.
Meyer’s software isn’t about to reveal a miraculous cure-all that can boost the immune system on demand. “People want a recipe to make COVID-19 go away, but that’s not how science works,” Beyaz says. “We don’t even know the molecular and cellular basis of complex diseases. We need to understand those processes and how they interact.” What Meyer can do—what she is doing—is provide the tools to figure out, at last, how those processes work. Slowly but surely, those tools are bringing us closer to the day when we can all be like that guy who forever beats the sniffles.
Lead image: Master1305/Shutterstock